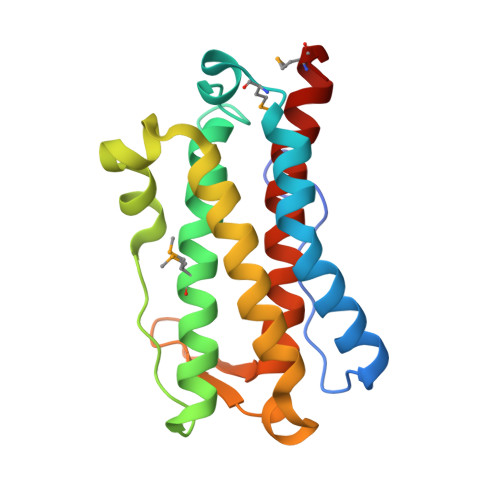
YfiT from Bacillus subtilis Is a Probable Metal-Dependent Hydrolase with an Unusual Four-Helix Bundle Topology
Rajan, S.S., Yang, X., Shuvalova, L., Collart, F., Anderson, W.F.(2004) Biochemistry 43: 15472-15479
- PubMed: 15581359
- DOI: https://doi.org/10.1021/bi048665r
- Primary Citation of Related Structures:
1RXQ - PubMed Abstract:
YfiT, a 19-kDa polypeptide from Bacillus subtilis, belongs to a small sequence family with members predominantly from Gram positive bacteria. We have determined the crystal structure of YfiT in complex with Ni(2+) to a resolution of 1.7 A. YfiT exists as a dimer and binds Ni(2+) in a 1:1 stoichiometry. The protein has an unusual four-helix bundle topology and coordinates Ni(2+) in an octahedral geometry with three conserved histidines and three waters. Although there is no similarity in their overall structures, the coordination geometry of the metal and the residues that constitute the putative active site in YfiT are similar to those of metalloproteases such as thermolysin. Our structural analyses suggest that YfiT might function as a metal-dependent hydrolase.
- Molecular Pharmacology and Biological Chemistry, Feinberg School of Medicine, Northwestern University, Chicago, Illinois 60611, USA.
Organizational Affiliation: