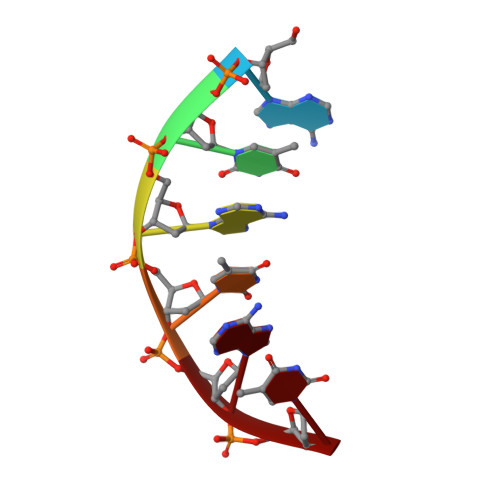
X-ray and NMR studies of the DNA oligomer d(ATATAT): Hoogsteen base pairing in duplex DNA.
Abrescia, N.G., Gonzalez, C., Gouyette, C., Subirana, J.A.(2004) Biochemistry 43: 4092-4100
- PubMed: 15065851
- DOI: https://doi.org/10.1021/bi0355140
- Primary Citation of Related Structures:
1RSB - PubMed Abstract:
We present and analyze the structure of the oligonucleotide d(ATATAT) found in two different forms by X-ray crystallography and in solution by NMR. We find that in both crystal lattices the oligonucleotide forms an antiparallel double helical duplex in which base pairing is of the Hoogsteen type. The double helix is apparently very similar to the standard B-form of DNA, with about 10 base pairs per turn. However, the adenines in the duplex are flipped over; as a result, the physicochemical features of both grooves of the helix are changed. In particular, the minor groove is narrow and hydrophobic. On the other hand, d(ATATAT) displays a propensity to adopt the B conformation in solution. These results confirm the polymorphism of AT-rich sequences in DNA. Furthermore, we show that extrahelical adenines and thymines can be minor groove binders in Hoogsteen DNA.
- Departament d'Enginyeria Química, Universitat Politècnica de Catalunya, Av. Diagonal 647, E-08028 Barcelona, Spain. nicola@strubi.ox.ac.uk
Organizational Affiliation: