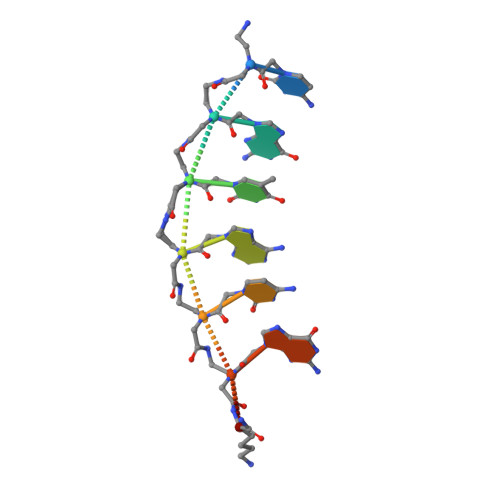
The Influence of a Chiral Amino Acid on the Helical Handedness of PNA in Solution and in Crystals
Rasmussen, H., Liljefors, T., Petersson, B., Nielsen, P.E., Kastrup, J.S.(2004) J Biomol Struct Dyn 21: 495-502
- PubMed: 14692794
- DOI: https://doi.org/10.1080/07391102.2004.10506943
- Primary Citation of Related Structures:
1RRU - PubMed Abstract:
The X-ray structure of a self-complementary PNA hexamer (H-CGTACG-L-Lys-NH(2)) has been determined to 2.35 A resolution. The introduction of an L-lysine moiety has previously been shown to induce a preferred left-handedness of the PNA double helices in aqueous solution. However, in the crystal structure an equal amount of interchanging right- and left-handed helices is observed. The lysine moieties are pointing into large solvent channels and no significant interactions between this moiety and the remaining PNA molecule are observed. In contrast, molecular mechanics calculations show a preference for the left-handed helix of this hexameric PNA in aqueous solution as expected. The calculations indicate that the difference in the free energy of solvation between the left-handed and the right-handed helix is the determining factor for the preference of the left-handed helix in aqueous solution.
- Department of Medicinal Chemistry, The Danish University of Pharmaceutical Sciences, Universitetsparken 2, DK-2100, Copenhagen, Denmark.
Organizational Affiliation: