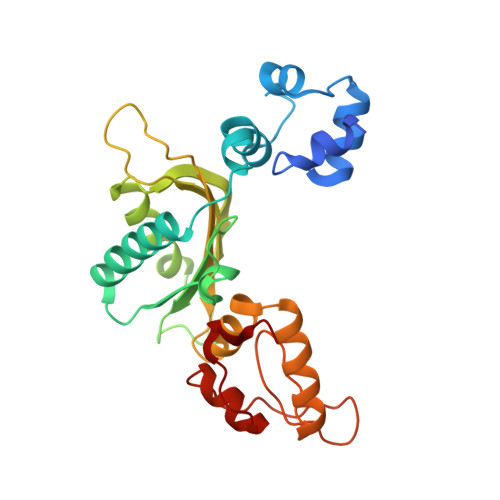
2.3 A crystal structure of the catalytic domain of DNA polymerase beta.
Davies 2nd., J.F., Almassy, R.J., Hostomska, Z., Ferre, R.A., Hostomsky, Z.(1994) Cell 76: 1123-1133
- PubMed: 8137427
- DOI: https://doi.org/10.1016/0092-8674(94)90388-3
- Primary Citation of Related Structures:
1RPL - PubMed Abstract:
The crystal structure of the catalytic domain of rat DNA polymerase beta (pol beta) has been determined at 2.3 A resolution and refined to an R factor of 0.22. The mixed alpha/beta protein has three subdomains arranged in an overall U shape reminiscent of other polymerase structures. The folding topology of pol beta, however, is unique. Two divalent metals bind near three aspartic acid residues implicated in the catalytic activity. In the presence of Mn2+ and dTTP, interpretable electron density is seen for two metals and the triphosphate, but not the deoxythymidine moiety. The principal interaction of the triphosphate moiety is with the bound divalent metals.
- Agouron Pharmaceuticals, Incorporated, San Diego, California 92121.
Organizational Affiliation: