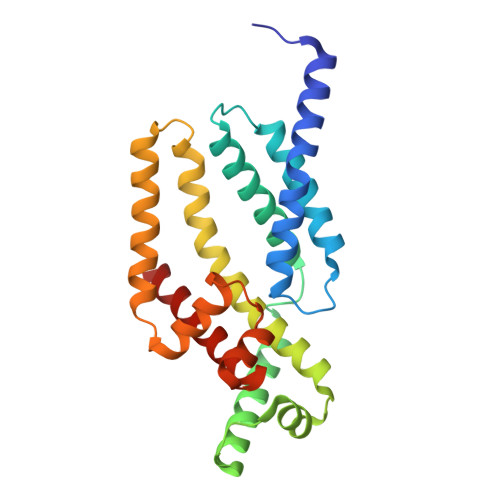
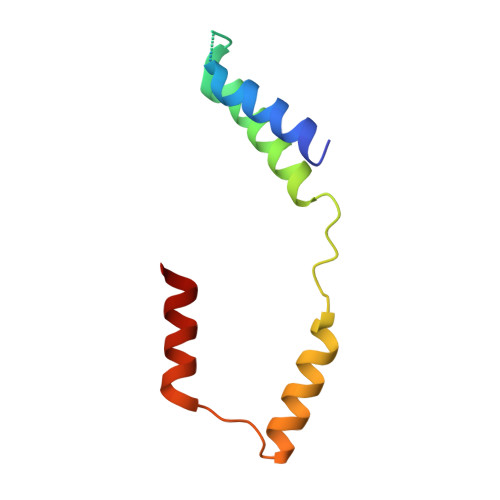
Crystal Structure of the Flagellar Sigma/Anti-Sigma Complex Sigma(28)/FlgM Reveals an Intact Sigma Factor in an Inactive Conformation
Sorenson, M.K., Ray, S.S., Darst, S.A.(2004) Mol Cell 14: 127-138
- PubMed: 15068809
- DOI: https://doi.org/10.1016/s1097-2765(04)00150-9
- Primary Citation of Related Structures:
1RP3, 1SC5 - PubMed Abstract:
The key regulators of bacterial transcription initiation are the sigma factors, which direct promoter recognition and melting but only after binding to the core RNA polymerase to form the holoenzyme. X-ray crystal structures of the flagellar sigma, sigma(28), in complex with its anti-sigma, FlgM, explain the inhibition mechanism of FlgM, including its ability to attack and destabilize the sigma(28)-holoenzyme. The sigma domains (sigma(2), sigma(3), and sigma(4)) pack together in a compact unit with extensive interdomain interfaces that bury the promoter binding determinants, including the -35 element recognition helix of sigma(4), which fits in an acidic groove on the surface of sigma(3). The structure illustrates the large rearrangements that sigma(28) must undergo to form the holoenzyme and provides insights into the regulation of sigma(28) promoter binding activity that may extend, at least in principle, to other sigmas.
- The Rockefeller University, 1230 York Avenue, New York, NY 10021, USA.
Organizational Affiliation: