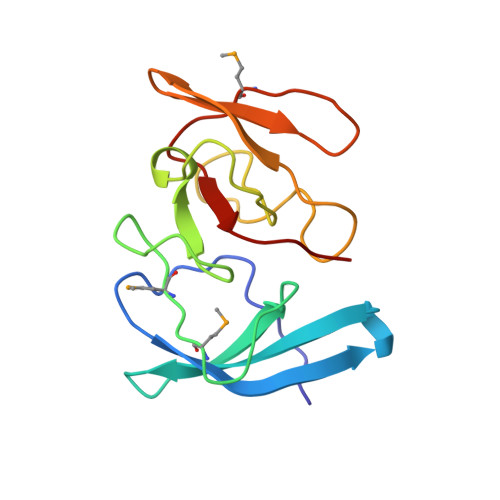
The three-dimensional structure of the RNA-binding domain of ribosomal protein L2; a protein at the peptidyl transferase center of the ribosome.
Nakagawa, A., Nakashima, T., Taniguchi, M., Hosaka, H., Kimura, M., Tanaka, I.(1999) EMBO J 18: 1459-1467
- PubMed: 10075918
- DOI: https://doi.org/10.1093/emboj/18.6.1459
- Primary Citation of Related Structures:
1RL2 - PubMed Abstract:
Ribosomal protein L2 is the largest protein component in the ribosome. It is located at or near the peptidyl transferase center and has been a prime candidate for the peptidyl transferase activity. It binds directly to 23S rRNA and plays a crucial role in its assembly. The three-dimensional structure of the RNA-binding domain of L2 from Bacillus stearothermophilus has been determined at 2.3 A resolution by X-ray crystallography using the selenomethionyl MAD method. The RNA-binding domain of L2 consists of two recurring motifs of approximately 70 residues each. The N-terminal domain (positions 60-130) is homologous to the OB-fold, and the C-terminal domain (positions 131-201) is homologous to the SH3-like barrel. Residues Arg86 and Arg155, which have been identified by mutation experiments to be involved in the 23S rRNA binding, are located at the gate of the interface region between the two domains. The molecular architecture suggests how this important protein has evolved from the ancient nucleic acid-binding proteins to create a 23S rRNA-binding domain in the very remote past.
- Division of Biological Sciences, Graduate School of Science, Hokkaido University, Sapporo 060-0810, Japan.
Organizational Affiliation: