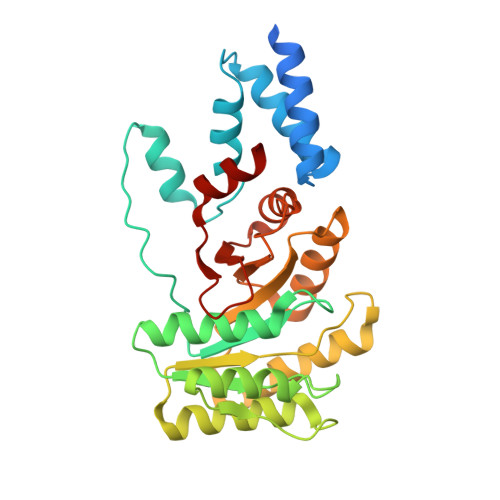
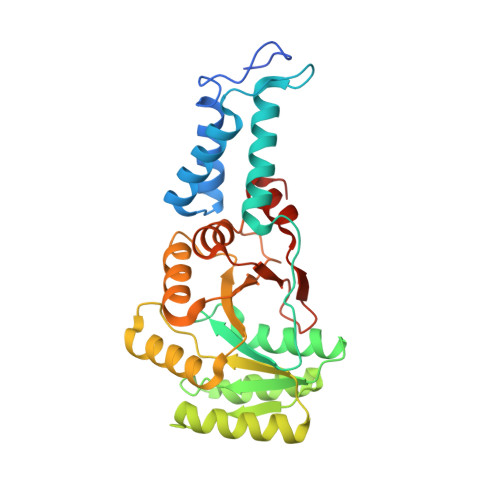
Substrate twinning activates the signal recognition particle and its receptor
Egea, P.F., Shan, S.O., Napetschnig, J., Savage, D.F., Walter, P., Stroud, R.M.(2004) Nature 427: 215-221
- PubMed: 14724630
- DOI: https://doi.org/10.1038/nature02250
- Primary Citation of Related Structures:
1RJ9 - PubMed Abstract:
Signal sequences target proteins for secretion from cells or for integration into cell membranes. As nascent proteins emerge from the ribosome, signal sequences are recognized by the signal recognition particle (SRP), which subsequently associates with its receptor (SR). In this complex, the SRP and SR stimulate each other's GTPase activity, and GTP hydrolysis ensures unidirectional targeting of cargo through a translocation pore in the membrane. To define the mechanism of reciprocal activation, we determined the 1.9 A structure of the complex formed between these two GTPases. The two partners form a quasi-two-fold symmetrical heterodimer. Biochemical analysis supports the importance of the extensive interaction surface. Complex formation aligns the two GTP molecules in a symmetrical, composite active site, and the 3'OH groups are essential for association, reciprocal activation and catalysis. This unique circle of twinned interactions is severed twice on hydrolysis, leading to complex dissociation after cargo delivery.
- Department of Biochemistry and Biophysics, University of California at San Francisco, California 94143-2240, USA. pascal@msg.ucsf.edu
Organizational Affiliation: