Structure of bovine liver rhodanese. I. Structure determination at 2.5 A resolution and a comparison of the conformation and sequence of its two domains.
Ploegman, J.H., Drent, G., Kalk, K.H., Hol, W.G.(1978) J Mol Biology 123: 557-594
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(1978) J Mol Biology 123: 557-594
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RHODANESE | 293 | Bos taurus | Mutation(s): 0 EC: 2.8.1.1 | 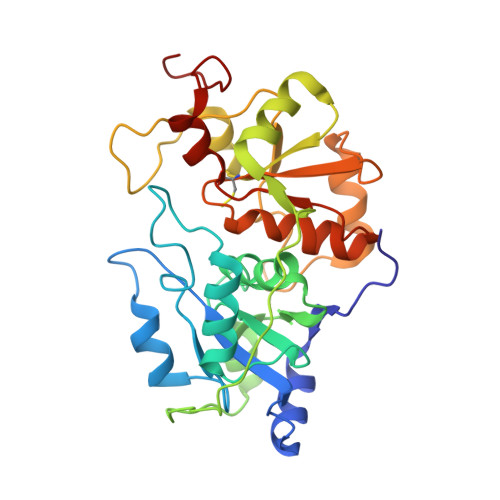 | |
UniProt | |||||
Find proteins for P00586 (Bos taurus) Explore P00586 Go to UniProtKB: P00586 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00586 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| CSS Query on CSS | A | L-PEPTIDE LINKING | C3 H7 N O2 S2 |  | CYS |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 156.1 | α = 90 |
| b = 49 | β = 98.6 |
| c = 42.2 | γ = 90 |