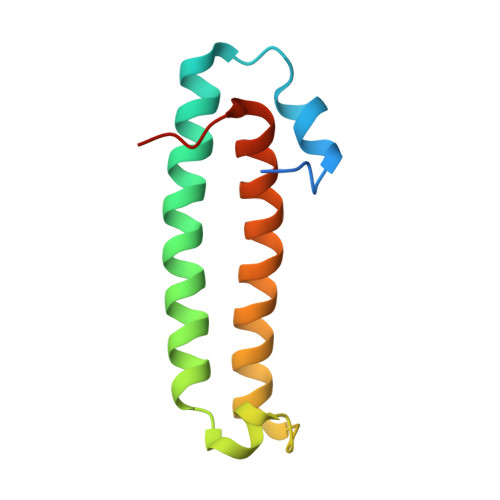
Quorum-sensing antiactivator TraM forms a dimer that dissociates to inhibit TraR
Chen, G., Malenkos, J.W., Cha, M.R., Fuqua, C., Chen, L.(2004) Mol Microbiol 52: 1641-1651
- PubMed: 15186414
- DOI: https://doi.org/10.1111/j.1365-2958.2004.04110.x
- Primary Citation of Related Structures:
1RFY - PubMed Abstract:
The quorum-sensing transcriptional activator TraR of Agrobacterium tumefaciens, which controls the replication and conjugal transfer of the tumour-inducing (Ti) virulence plasmid, is inhibited by the TraM antiactivator. The crystal structure of TraM reveals this protein to form a homodimer in which the monomer primarily consists of two long coiled alpha-helices, and one of the helices from each monomer also bundles to form the dimeric interface. The importance of dimerization is addressed by mutational studies in which disruption of the hydrophobic dimer interface leads to aggregation of TraM. Biochemical studies confirm that TraM exists as a homodimer in solution in equilibrium with the monomeric form, and also establish that the TraM-TraR complex is a heterodimer. Thus, the TraM homodimer undergoes dissociation in forming the antiactivation complex. Combined with the structure of TraR (Zhang et al., 2002, Nature 417: 971-974; Vannini et al., 2002, EMBO J 21: 4393-4401), our structural analysis suggests overlapping interactive surfaces in homodimeric TraM with those in the TraM-TraR complex and a mechanism for TraM inhibition on TraR.
- Department of Biology, Indiana University, 915 E. 3rd St., Bloomington, IN 47405, USA.
Organizational Affiliation: