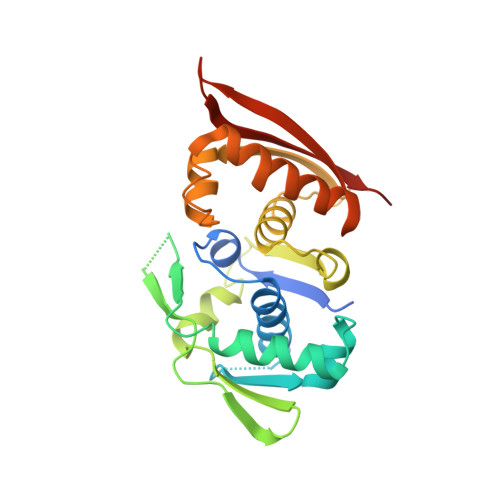
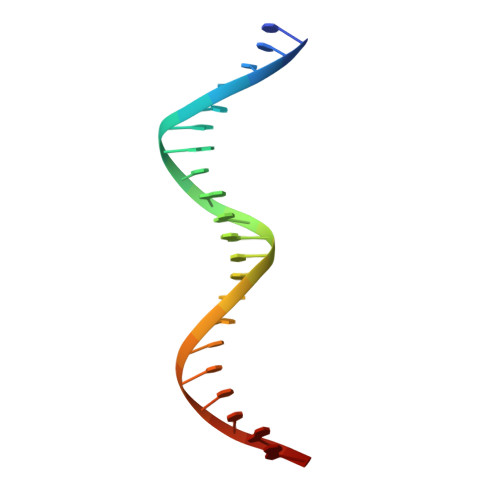
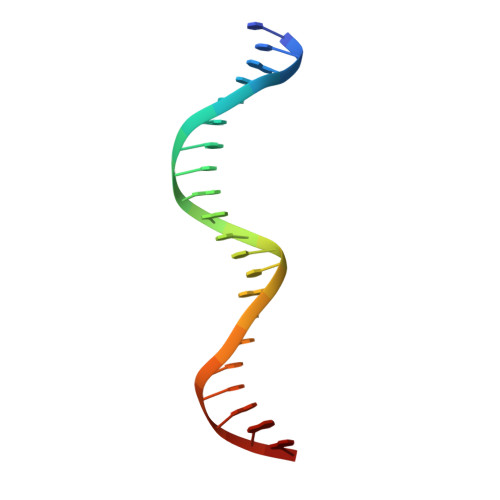
Crystal structure of a prokaryotic replication initiator protein bound to DNA at 2.6 A resolution.
Komori, H., Matsunaga, F., Higuchi, Y., Ishiai, M., Wada, C., Miki, K.(1999) EMBO J 18: 4597-4607
- PubMed: 10469640
- DOI: https://doi.org/10.1093/emboj/18.17.4597
- Primary Citation of Related Structures:
1REP - PubMed Abstract:
The initiator protein (RepE) of F factor, a plasmid involved in sexual conjugation in Escherichia coli, has dual functions during the initiation of DNA replication which are determined by whether it exists as a dimer or as a monomer. A RepE monomer functions as a replication initiator, but a RepE dimer functions as an autogenous repressor. We have solved the crystal structure of the RepE monomer bound to an iteron DNA sequence of the replication origin of plasmid F. The RepE monomer consists of topologically similar N- and C-terminal domains related to each other by internal pseudo 2-fold symmetry, despite the lack of amino acid similarities between the domains. Both domains bind to the two major grooves of the iteron (19 bp) with different binding affinities. The C-terminal domain plays the leading role in this binding, while the N-terminal domain has an additional role in RepE dimerization. The structure also suggests that superhelical DNA induced at the origin of plasmid F by four RepEs and one HU dimer has an essential role in the initiation of DNA replication.
- Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, USA.
Organizational Affiliation: