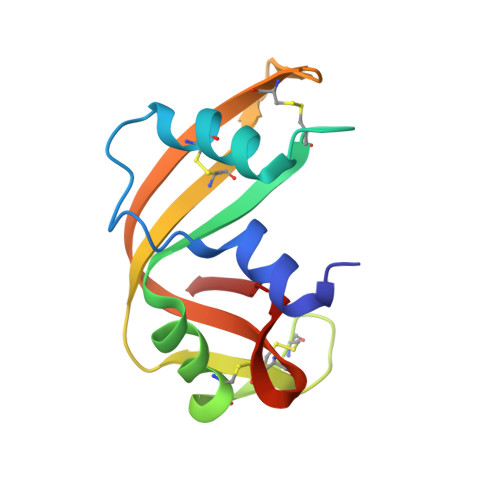
Structure of ribonuclease A derivative II at 2.1-A resolution.
Boque, L., Gracia Coll, M., Vilanova, M., Cuchillo, C.M., Fita, I.(1994) J Biological Chem 269: 19707-19712
- PubMed: 8051049
- Primary Citation of Related Structures:
1RBN - PubMed Abstract:
The crystal structure of bovine pancreatic ribonuclease A derivative II, a covalent derivative obtained by reaction of 6-chloropurine 9-beta-D-ribofuranosyl 5'-monophosphate with the alpha-amino group of Lys-1, has been determined and refined at 2.1-A resolution with an agreement factor R = 0.166 for 6254 reflections in the resolution shell 8.0 to 2.1 A. Crystals are orthorhombic and belong to space group C222(1) with unit cell parameters a = 75.73 A, b = 57.85 A, and c = 53.26 A. This crystal packing had never been reported before for pancreatic ribonuclease nor its complexes. The structure found is in accordance with the location of p2, B3, and R3 subsites at the N-terminal region of the protein and provides an explanation of the catalytic behavior observed for this derivative. In particular, differences in kinetic parameters and in the pKa value of His-119 between derivative II and native ribonuclease A can be interpreted on the basis of the position of the phosphate moiety within the derivative structure. Some uncertainty remains on the nucleotide sugar conformation determined.
- Departament d'Enginyeria Química, Universitat Politècnica de Catalunya, Barcelona, Spain.
Organizational Affiliation: