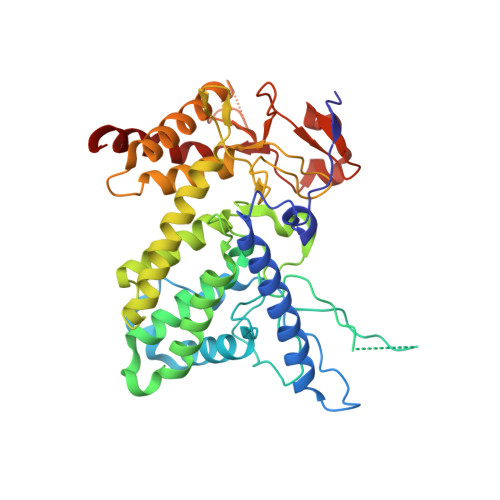
Azospirillum irakense pectate lyase displays a toroidal fold.
Novoa De Armas, H., Verboven, C., De Ranter, C., Desair, J., Vande Broek, A., Vanderleyden, J., Rabijns, A.(2004) Acta Crystallogr D Biol Crystallogr 60: 999-1007
- PubMed: 15159558
- DOI: https://doi.org/10.1107/S090744490400602X
- Primary Citation of Related Structures:
1R76 - PubMed Abstract:
The three-dimensional structure of Azospirillum irakense pectate lyase (PelA) has been determined at a resolution of 2.65 A. The crystals are hexagonal, belonging to space group P6(5)22, with unit-cell parameters a = b = 85.37, c = 231.32 angstroms. Phase information was derived from a multiple-wavelength anomalous dispersion (MAD) experiment using a Hg derivative. Refinement of the model converged to Rcryst = 20.08% and Rfree = 25.87%. The overall structure of PelA does not adopt the characteristic parallel beta-helix fold displayed by pectate lyases from polysaccharide lyase (PL) families PL1, PL3 and PL9. Instead, it displays a predominantly alpha-helical structure with irregular coils and short beta-strands, similar to the recently reported structure of the catalytic module of the Cellvibrio japonicus pectate lyase Pel10Acm. The topologies of the two structures have been compared. They show two 'domains' with the interface between them being a wide-open central groove in which the active site is located. The active sites of the crystal structures are also compared and their similarities and differences are discussed.
- Laboratorium voor Analytische Chemie en Medicinale Fysicochemie, Faculteit Farmaceutische Wetenschappen, K.U. Leuven, E. Van Evenstraat 4, B-3000 Leuven, Belgium.
Organizational Affiliation: