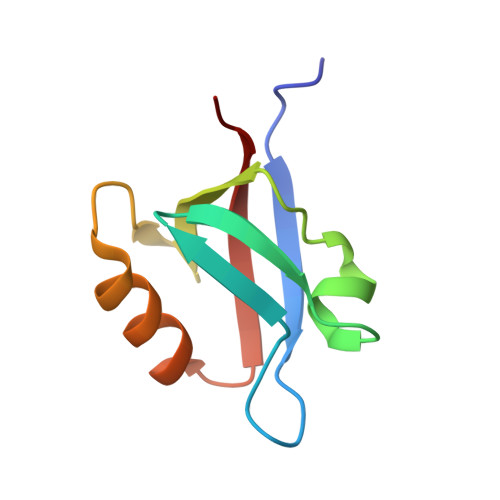
The PDZ2 domain of syntenin at ultra-high resolution: bridging the gap between small molecule and macromolecular crystal chemistry
Kang, B.S., Devedjiev, Y., Derewenda, U., Derewenda, Z.S.(2004) J Mol Biology 338: 483-493
- PubMed: 15081807
- DOI: https://doi.org/10.1016/j.jmb.2004.02.057
- Primary Citation of Related Structures:
1R6J - PubMed Abstract:
The crystal structure of the second PDZ domain of the scaffolding protein syntenin was solved using data extending to 0.73 A resolution. The crystallographic model, including the hydrogen atoms and the anisotropic displacement parameters, was refined to a conventional R-factor of 7.5% and Rfree of 8.7%, making it the most precise crystallographic model of a protein molecule to date. The model reveals discrete disorder in several places in the molecule, and significant plasticity of the peptide bond, with some omega angles deviating by nearly 20 degrees from planarity. Most hydrogen atoms are easily identifiable in the electron density and weak hydrogen bonds of the C-H...O type are clearly visible between the beta-strands. The study sets a new standard for high-resolution protein crystallography.
- Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA 22908-0736, USA.
Organizational Affiliation: