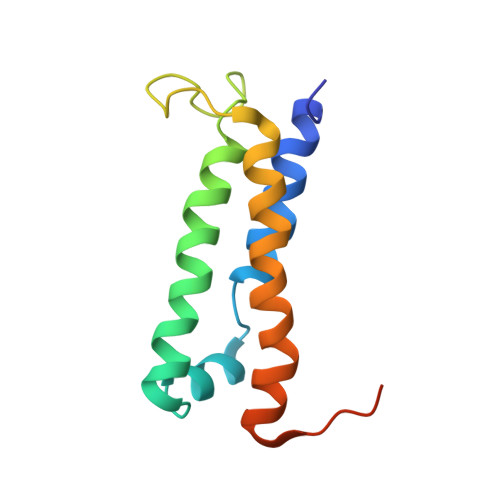
The solution structure of type III effector protein AvrPto reveals conformational and dynamic features important for plant pathogenesis.
Wulf, J., Pascuzzi, P.E., Fahmy, A., Martin, G.B., Nicholson, L.K.(2004) Structure 12: 1257-1268
- PubMed: 15242602
- DOI: https://doi.org/10.1016/j.str.2004.04.017
- Primary Citation of Related Structures:
1R5E - PubMed Abstract:
Pseudomonas syringae pv. tomato, the causative agent of bacterial speck disease of tomato, uses a type III secretion system (TTSS) to deliver effector proteins into the host cell. In resistant plants, the bacterial effector protein AvrPto physically interacts with the host Pto kinase and elicits antibacterial defense responses. In susceptible plants, which lack the Pto kinase, AvrPto acts as a virulence factor to promote bacterial growth. The solution structure of AvrPto reveals a functional core consisting of a three-helix bundle motif flanked by disordered N- and C-terminal tails. Residues required for Pto binding lie in a 19 residue Omega loop. Modeling suggests a hydrophobic patch involving the activation loop of Pto forms a contact surface with the AvrPto Omega loop and that helix packing mediates interactions between AvrPto and putative virulence targets Api2 and Api3. The AvrPto structure has a low stability that may facilitate chaperone-independent secretion by the TTSS.
- Department of Molecular Biology and Genetics, Cornell University, Ithaca, NY 14853, USA.
Organizational Affiliation: