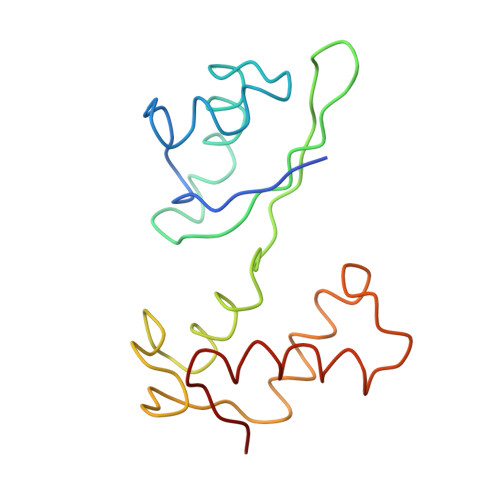
Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron Microscopy
Valle, M., Zavialov, A., Li, W., Stagg, S.M., Sengupta, J., Nielsen, R.C., Nissen, P., Harvey, S.C., Ehrenberg, M., Frank, J.(2003) Nat Struct Biol 10: 899-906
- PubMed: 14566331
- DOI: https://doi.org/10.1038/nsb1003
- Primary Citation of Related Structures:
1QZA, 1QZB, 1QZC, 1QZD, 1R2W, 1R2X - PubMed Abstract:
Aminoacyl-tRNAs (aa-tRNAs) are delivered to the ribosome as part of the ternary complex of aa-tRNA, elongation factor Tu (EF-Tu) and GTP. Here, we present a cryo-electron microscopy (cryo-EM) study, at a resolution of approximately 9 A, showing that during the incorporation of the aa-tRNA into the 70S ribosome of Escherichia coli, the flexibility of aa-tRNA allows the initial codon recognition and its accommodation into the ribosomal A site. In addition, a conformational change observed in the GTPase-associated center (GAC) of the ribosomal 50S subunit may provide the mechanism by which the ribosome promotes a relative movement of the aa-tRNA with respect to EF-Tu. This relative rearrangement seems to facilitate codon recognition by the incoming aa-tRNA, and to provide the codon-anticodon recognition-dependent signal for the GTPase activity of EF-Tu. From these new findings we propose a mechanism that can explain the sequence of events during the decoding of mRNA on the ribosome.
- Howard Hughes Medical Institute, Health Research, Inc. at the Wadsworth Center, Empire State Plaza, Albany, New York 12201-0509, USA.
Organizational Affiliation: