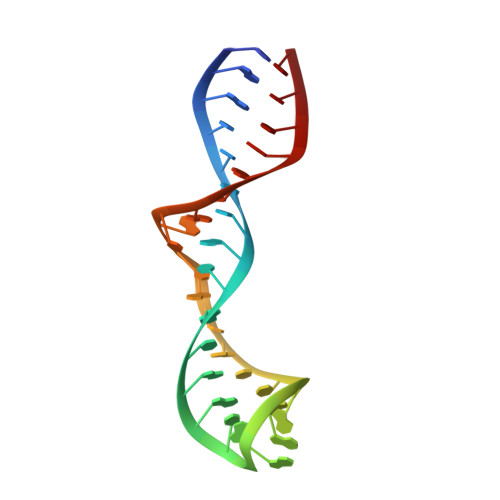
Solution structure of domain 5 of a group II intron ribozyme reveals a new RNA motif.
Sigel, R.K., Sashital, D.G., Abramovitz, D.L., Palmer, A.G., Butcher, S.E., Pyle, A.M.(2004) Nat Struct Mol Biol 11: 187-192
- PubMed: 14745440
- DOI: https://doi.org/10.1038/nsmb717
- Primary Citation of Related Structures:
1R2P - PubMed Abstract:
Domain 5 (D5) is the central core of group II intron ribozymes. Many base and backbone substituents of this highly conserved hairpin participate in catalysis and are crucial for binding to other intron domains. We report the solution structures of the 34-nucleotide D5 hairpin from the group II intron ai5 gamma in the absence and presence of divalent metal ions. The bulge region of D5 adopts a novel fold, where G26 adopts a syn conformation and flips down into the major groove of helix 1, close to the major groove face of the catalytic AGC triad. The backbone near G26 is kinked, exposing the base plane of the adjacent A-U pair to the solvent and causing bases of the bulge to stack intercalatively. Metal ion titrations reveal strong Mg(2+) binding to a minor groove shelf in the D5 bulge. Another distinct metal ion-binding site is observed along the minor groove side of the catalytic triad, in a manner consistent with metal ion binding in the ribozyme active site.
- Department of Chemistry, Winterthurerstrasse 190, University of Zürich, CH-8057 Zürich, Switzerland.
Organizational Affiliation: