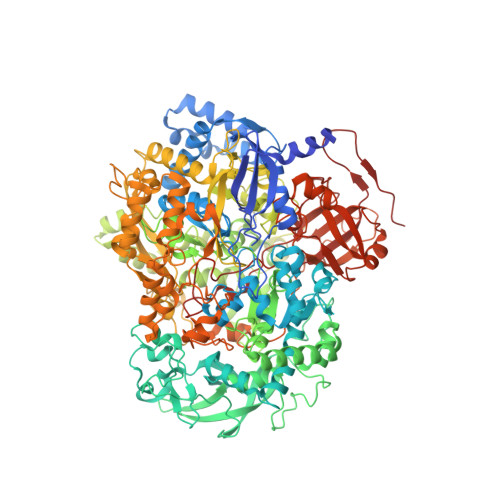
Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes
Jormakka, M., Richardson, D., Byrne, B., Iwata, S.(2004) Structure 12: 95-104
- PubMed: 14725769
- DOI: https://doi.org/10.1016/j.str.2003.11.020
- Primary Citation of Related Structures:
1R27 - PubMed Abstract:
The structure of the catalytic and electron-transfer subunits (NarGH) of the integral membrane protein, respiratory nitrate reductase (Nar) has been determined to 2.0 A resolution revealing the molecular architecture of this Mo-bisMGD (molybdopterin-guanine-dinucleotide) containing enzyme which includes a previously undetected FeS cluster. Nar, together with the related enzyme formate dehydrogenase (Fdh-N), is a key enzyme in the generation of proton motive force across the membrane in Escherichia coli nitrate respiration. A comparative study revealed that Nar and Fdh-N employ different approaches for acquiring substrate, reflecting different catalytic mechanisms. Nar uses a very narrow and nonpolar substrate-conducting cavity with a nonspecific substrate binding site, whereas Fdh-N accommodates a wider, positively charged substrate-conducting cavity with a more specific substrate binding site. The Nar structure also demonstrates the first example of an Asp side chain acting as a Mo ligand providing a structural basis for the classification of Mo-bisMGD enzymes.
- Division of Biomedical Sciences, Imperial College London, SW7 2AZ, UK.
Organizational Affiliation: