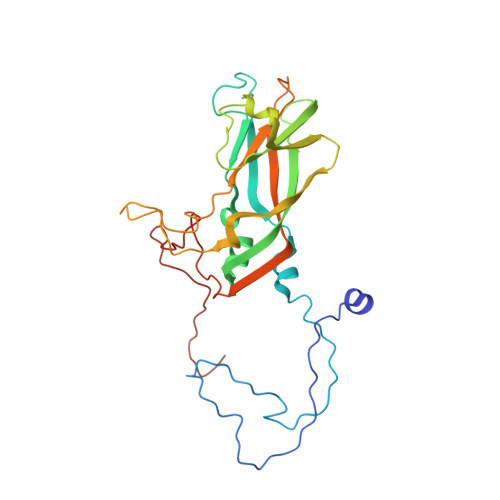
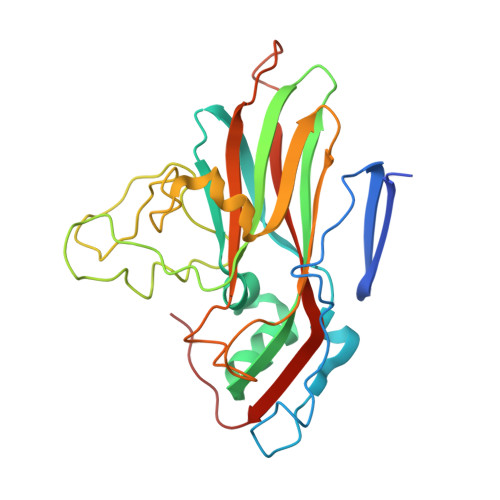
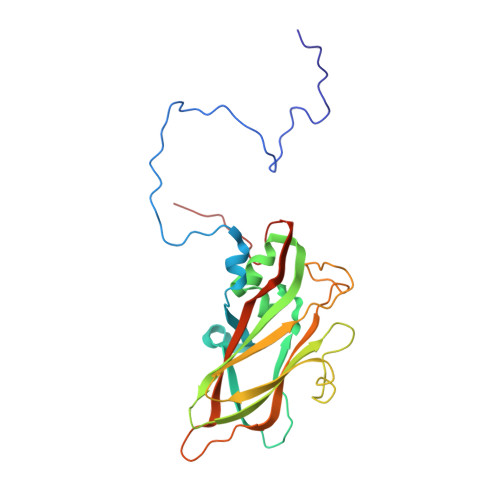
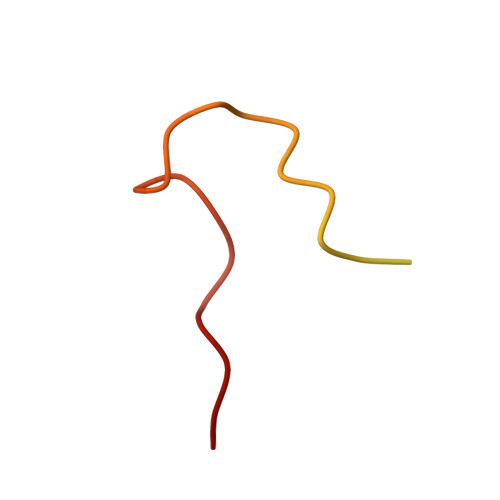
Crystal structure of human rhinovirus serotype 1A (HRV1A).
Kim, S.S., Smith, T.J., Chapman, M.S., Rossmann, M.G., Pevear, D.C., Dutko, F.J., Felock, P.J., Diana, G.D., McKinlay, M.A.(1989) J Mol Biology 210: 91-111
- PubMed: 2555523
- DOI: https://doi.org/10.1016/0022-2836(89)90293-3
- Primary Citation of Related Structures:
1R1A - PubMed Abstract:
The structure of human rhinovirus 1A (HRV1A) has been determined to 3.2 A resolution using phase refinement and extension by symmetry averaging starting with phases at 5 A resolution calculated from the known human rhinovirus 14 (HRV14) structure. The polypeptide backbone structures of HRV1A and HRV14 are similar, but the exposed surfaces are rather different. Differential charge distribution of amino acid residues in the "canyon", the putative receptor binding site, provides a possible explanation for the difference in minor versus major receptor group specificities, represented by HRV1A and HRV14, respectively. The hydrophobic pocket in VP1, into which antiviral compounds bind, is in an "open" conformation similar to that observed in drug-bound HRV14. Drug binding in HRV1A does not induce extensive conformational changes, in contrast to the case of HRV14.
- Department of Biological Sciences, Purdue University, West Lafayette, IN 47907.
Organizational Affiliation: