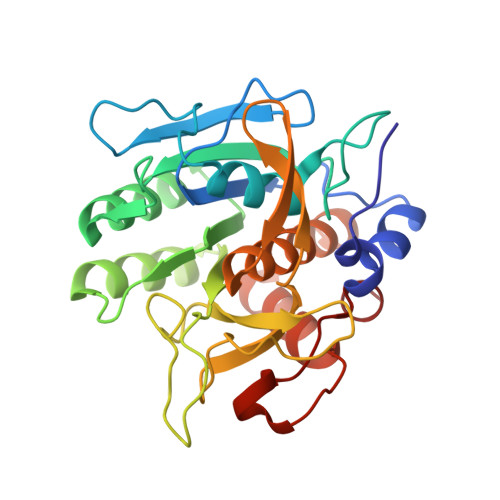
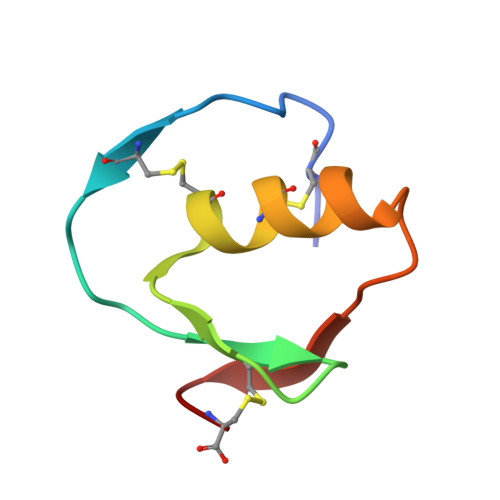
Structure and energetics of protein-protein interactions: the role of conformational heterogeneity in OMTKY3 binding to serine proteases
Horn, J.R., Ramaswamy, S., Murphy, K.P.(2003) J Mol Biology 331: 497-508
- PubMed: 12888355
- DOI: https://doi.org/10.1016/s0022-2836(03)00783-6
- Primary Citation of Related Structures:
1R0R - PubMed Abstract:
Proteins with flexible binding surfaces can interact with numerous binding partners. However, this promiscuity is more difficult to understand in "rigid-body" proteins, whose binding results in little, or no, change in the position of backbone atoms. The binding of Kazal inhibitors to serine proteases is considered a classic case of rigid-body binding, although they bind to a wide range of proteases. We have studied the thermodynamics of binding of the Kazal serine protease inhibitor, turkey ovomucoid third domain (OMTKY3), to the serine protease subtilisin Carlsberg using isothermal titration calorimetry and have determined the crystal structure of the complex at very high resolution (1.1A). Comparison of the binding energetics and structure to other OMTKY3 interactions demonstrates that small changes in the position of side-chains can make significant contributions to the binding thermodynamics, including the enthalpy of binding. These effects emphasize that small, "rigid-body" proteins are still dynamic structures, and these dynamics make contributions to both the enthalpy and entropy of binding interactions.
- Department of Biochemistry, College of Medicine, University of Iowa, Iowa City, IA 52242, USA.
Organizational Affiliation: