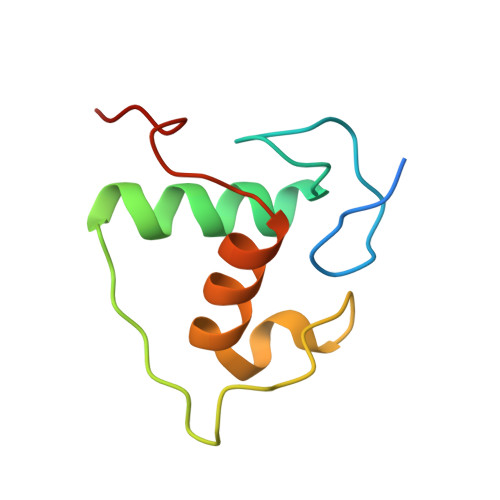
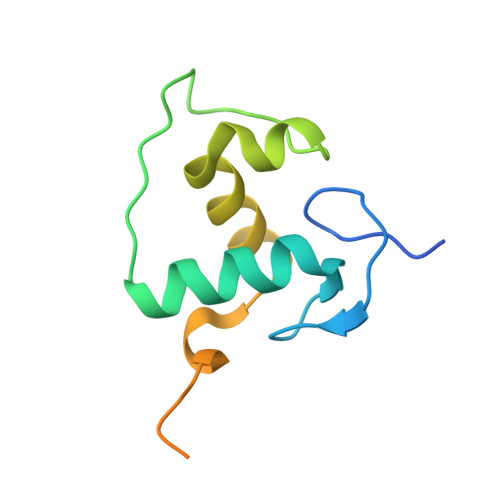
Structure of the Heterodimeric Ecdysone Receptor DNA-binding Complex
Devarakonda, S., Harp, J.M., Kim, Y., Ozyhar, A., Rastinejad, F.(2003) EMBO J 22: 5827-5840
- PubMed: 14592980
- DOI: https://doi.org/10.1093/emboj/cdg569
- Primary Citation of Related Structures:
1R0N, 1R0O - PubMed Abstract:
Ecdysteroids initiate molting and metamorphosis in insects via a heterodimeric receptor consisting of the ecdysone receptor (EcR) and ultraspiracle (USP). The EcR-USP heterodimer preferentially mediates transcription through highly degenerate pseudo-palindromic response elements, resembling inverted repeats of 5'-AGGTCA-3' separated by 1 bp (IR-1). The requirement for a heterodimeric arrangement of EcR-USP subunits to bind to a symmetric DNA is unusual within the nuclear receptor superfamily. We describe the 2.24 A structure of the EcR-USP DNA-binding domain (DBD) heterodimer bound to an idealized IR-1 element. EcR and USP use similar surfaces, and rely on the deformed minor groove of the DNA to establish protein-protein contacts. As retinoid X receptor (RXR) is the mammalian homolog of USP, we also solved the 2.60 A crystal structure of the EcR-RXR DBD heterodimer on IR-1 and found the dimerization and DNA-binding interfaces to be the same as in the EcR-USP complex. Sequence alignments indicate that the EcR-RXR heterodimer is an important model for understanding how the FXR-RXR heterodimer binds to IR-1 sites.
- Department of Pharmacology, University of Virginia Health System, Charlottesville, VA 22908, USA.
Organizational Affiliation: