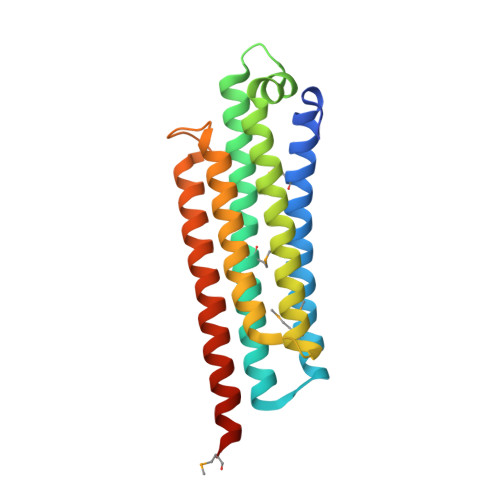
Structural definition of the F-actin-binding THATCH domain from HIP1R
Brett, T.J., Legendre-Guillemin, V., McPherson, P.S., Fremont, D.H.(2006) Nat Struct Mol Biol 13: 121-130
- PubMed: 16415883
- DOI: https://doi.org/10.1038/nsmb1043
- Primary Citation of Related Structures:
1R0D - PubMed Abstract:
Huntingtin-interacting protein-1 related (HIP1R) has a crucial protein-trafficking role, mediating associations between actin and clathrin-coated structures at the plasma membrane and trans-Golgi network. Here, we characterize the F-actin-binding region of HIP1R, termed the talin-HIP1/R/Sla2p actin-tethering C-terminal homology (THATCH) domain. The 1.9-A crystal structure of the human HIP1R THATCH core reveals a large sequence-conserved surface patch created primarily by residues from the third and fourth helices of a unique five-helix bundle. Point mutations of seven contiguous patch residues produced significant decreases in F-actin binding. We also show that THATCH domains have a conserved C-terminal latch capable of oligomerizing the core, thereby modulating F-actin engagement. Collectively, these results establish a framework for investigating the links between endocytosis and actin dynamics mediated by THATCH domain-containing proteins.
- Department of Pathology and Immunology, Washington University School of Medicine, 660 S. Euclid Ave, St. Louis, Missouri 63110, USA.
Organizational Affiliation: