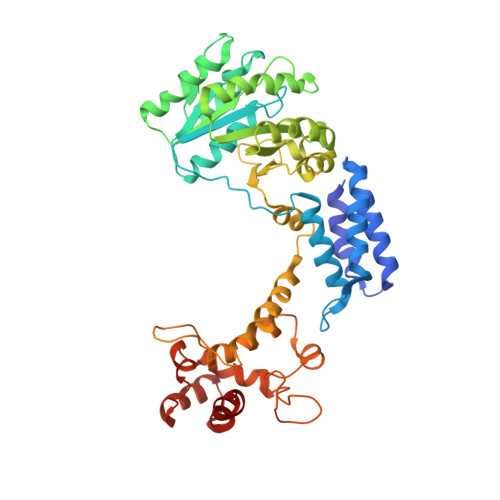
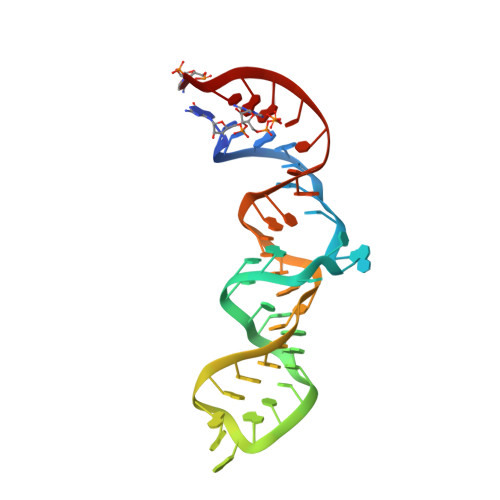
Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication
Rosendal, K.R., Wild, K., Montoya, G., Sinning, I.(2003) Proc Natl Acad Sci U S A 100: 14701-14706
- PubMed: 14657338
- DOI: https://doi.org/10.1073/pnas.2436132100
- Primary Citation of Related Structures:
1QZW, 1QZX - PubMed Abstract:
Targeting of secretory and membrane proteins by the signal recognition particle (SRP) is evolutionarily conserved, and the multidomain protein SRP54 acts as the key player in SRP-mediated protein transport. Binding of a signal peptide to SRP54 at the ribosome is coordinated with GTP binding and subsequent complex formation with the SRP receptor. Because these functions are localized to distinct domains of SRP54, communication between them is essential. We report the crystal structures of SRP54 from the Archaeon Sulfolobus solfataricus with and without its cognate SRP RNA binding site (helix 8) at 4-A resolution. The two structures show the flexibility of the SRP core and the position of SRP54 relative to the RNA. A long linker helix connects the GTPase (G domain) with the signal peptide binding (M) domain, and a hydrophobic contact between the N and M domains relates the signal peptide binding site to the G domain. Hinge regions are identified in the linker between the G and M domains (292-LGMGD) and in the N-terminal part of the M domain, which allow for structural rearrangements within SRP54 upon signal peptide binding at the ribosome.
- Biochemie-Zentrum Heidelberg, INF 328, D-69120 Heidelberg, Germany.
Organizational Affiliation: