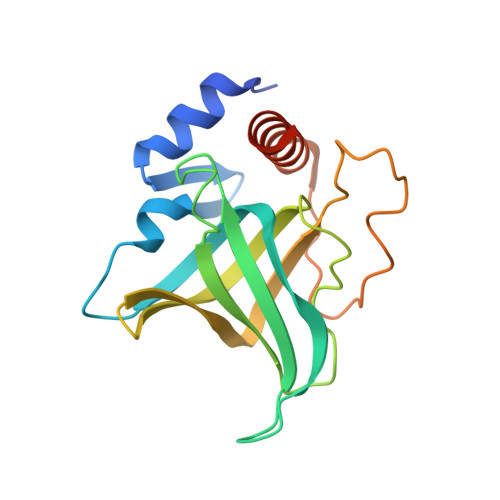
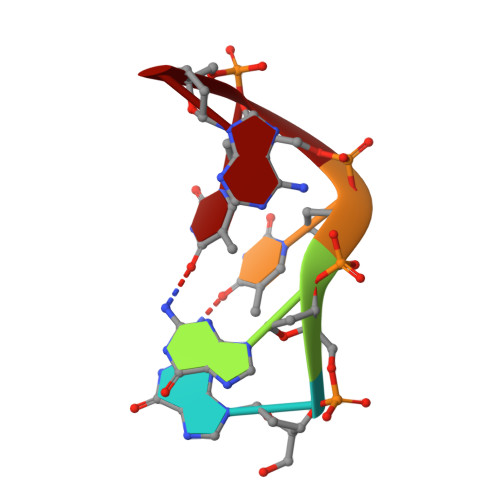
DNA self-recognition in the structure of Pot1 bound to telomeric single-stranded DNA
Lei, M., Podell, E.R., Baumann, P., Cech, T.R.(2003) Nature 426: 198-203
- PubMed: 14614509
- DOI: https://doi.org/10.1038/nature02092
- Primary Citation of Related Structures:
1QZG, 1QZH - PubMed Abstract:
Telomeres, specialized protein-DNA complexes that cap the ends of linear chromosomes, are essential for protecting chromosomes from degradation and end-to-end fusions. The Pot1 (protection of telomeres 1) protein is a widely distributed eukaryotic end-capping protein, having been identified in fission yeast, microsporidia, plants and animals. Schizosaccharomyces pombe Pot1p is essential for telomere maintenance, and human POT1 has been implicated in telomerase regulation. Pot1 binds telomeric single-stranded DNA (ssDNA) with exceptionally high sequence specificity, the molecular basis of which has been unknown. Here we describe the 1.9-A-resolution crystal structure of the amino-terminal DNA-binding domain of S. pombe Pot1p complexed with ssDNA. The protein adopts an oligonucleotide/oligosaccharide-binding (OB) fold with two loops that protrude to form a clamp for ssDNA binding. The structure explains the sequence specificity of binding: in the context of the Pot1 protein, DNA self-recognition involving base-stacking and unusual G-T base pairs compacts the DNA. Any sequence change disrupts the ability of the DNA to form this structure, preventing it from contacting the array of protein hydrogen-bonding groups. The structure also explains how Pot1p avoids binding the vast excess of RNA in the nucleus.
- Howard Hughes Medical Institute, Department of Chemistry and Biochemistry, University of Colorado, Boulder, Colorado 80309-0215, USA.
Organizational Affiliation: