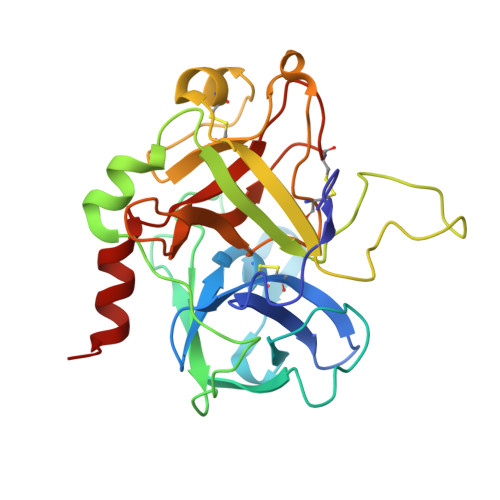
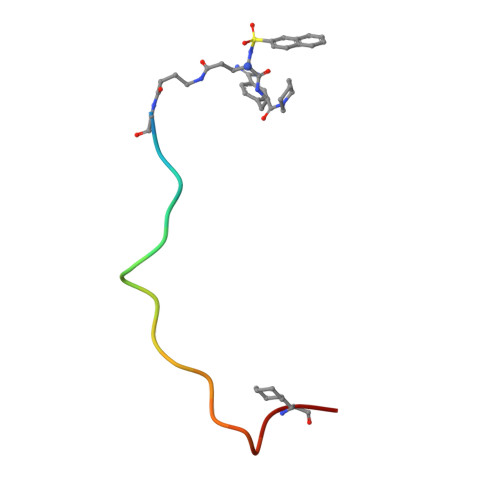
Design and evaluation of novel bivalent thrombin inhibitors based on amidinophenylalanines.
Steinmetzer, T., Renatus, M., Kunzel, S., Eichinger, A., Bode, W., Wikstrom, P., Hauptmann, J., Sturzebecher, J.(1999) Eur J Biochem 265: 598-605
- PubMed: 10504391
- DOI: https://doi.org/10.1046/j.1432-1327.1999.00742.x
- Primary Citation of Related Structures:
1QUR - PubMed Abstract:
Two bivalent thrombin inhibitors were synthesized, which consist of a benzamidine-based active-site-blocking segment, a fibrinogen recognition exosite inhibitor and a peptidic linker connecting these fragments. BZA-1 hirulog contains an Nalpha-(2-naphthylsulfonyl)-S-3-amidinophenylalanyl-is onipecotic acid residue connected via the carboxyl group to the linker segment. The active-site-directed moiety of BZA-2 hirulog [Nalpha-(2-naphthylsulfonyl-glutamyl)-R-4-amidinophenylal anyl-piperid ide] was coupled to the linker via the side chain of the glutamic acid. Both BZA-hirulogs contain almost identical linker-exo site inhibitor parts, except for the substitution of a glycine as the first linker residue in BZA-1 hirulog by a gamma-amino butyric acid in BZA-2 hirulog, thus increasing flexibility and linker length by two additional atoms. BZA-1 hirulog showed moderate potency (Ki = 0. 50 +/- 0.14 nM), while BZA-2 hirulog was characterized as a slow, tight binding inhibitor of thrombin (Ki = 0.29 +/- 0.08 pM). The stability in human plasma of both analogs was strongly improved compared with hirulog-1. For BZA-2 hirulog a significantly reduced plasma clearance was observed after intravenous injection in rats compared with BZA-1 hirulog and hirulog-1. The X-ray structure of the BZA-2 hirulog in complex with human alpha-thrombin was solved and confirmed the expected bivalent binding mode.
- Friedrich-Schiller-Universität, Institut für Biochemie & Biophysik, Jena, Germany. steinmetzer@merlin.biologie.uni-jena.de
Organizational Affiliation: