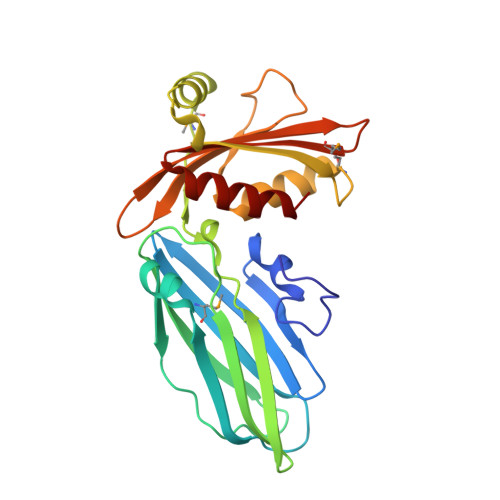
Crystal structure of the alpha appendage of AP-2 reveals a recruitment platform for clathrin-coat assembly.
Traub, L.M., Downs, M.A., Westrich, J.L., Fremont, D.H.(1999) Proc Natl Acad Sci U S A 96: 8907-8912
- PubMed: 10430869
- DOI: https://doi.org/10.1073/pnas.96.16.8907
- Primary Citation of Related Structures:
1QTP, 1QTS - PubMed Abstract:
AP-2 adaptors regulate clathrin-bud formation at the cell surface by recruiting clathrin trimers to the plasma membrane and by selecting certain membrane proteins for inclusion within the developing clathrin-coat structure. These functions are performed by discrete subunits of the adaptor heterotetramer. The carboxyl-terminal appendage of the AP-2 alpha subunit appears to regulate the translocation of several endocytic accessory proteins to the bud site. We have determined the crystal structure of the alpha appendage at 1.4-A resolution by multiwavelength anomalous diffraction phasing. It is composed of two distinct structural modules, a beta-sandwich domain and a mixed alpha-beta platform domain. Structure-based mutagenesis shows that alterations to the molecular surface of a highly conserved region on the platform domain differentially affect associations of the appendage with amphiphysin, eps15, epsin, and AP180, revealing a common protein-binding interface.
- Department of Internal Medicine, Washington University School of Medicine, 660 South Euclid Avenue, St. Louis, MO 63110, USA.
Organizational Affiliation: