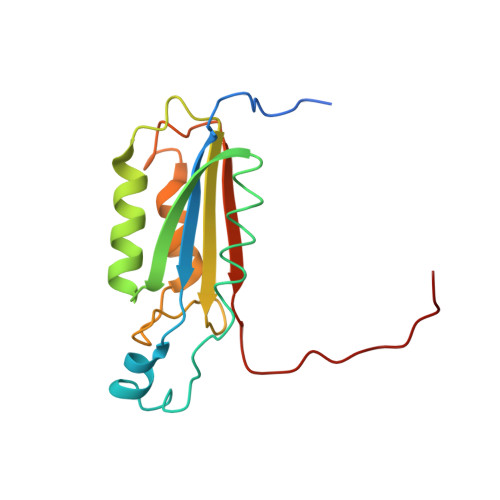
The atomic-resolution structure of human caspase-8, a key activator of apoptosis.
Watt, W., Koeplinger, K.A., Mildner, A.M., Heinrikson, R.L., Tomasselli, A.G., Watenpaugh, K.D.(1999) Structure 7: 1135-1143
- PubMed: 10508785
- DOI: https://doi.org/10.1016/s0969-2126(99)80180-4
- Primary Citation of Related Structures:
1QTN - PubMed Abstract:
Caspases are a family of cysteine proteases that have important intracellular roles in inflammation and apoptosis. Caspase-8 activates downstream caspases which are unable to carry out autocatalytic processing and activation. Caspase-8 is designated as an initiator caspase and is believed to sit at the apex of the Fas- or TNF-mediated apoptotic cascade. In view of this role, the enzyme is an attractive target for the design of inhibitors aimed at blocking the undesirable cell death associated with a range of degenerative disorders. The structure of recombinant human caspase-8, covalently modified with the inhibitor acetyl-Ile-Glu-Thr-Asp-aldehyde, has been determined by X-ray crystallography to 1.2 A resolution. The asymmetric unit contains the p18-p11 heterodimer; the biologically important molecule contains two dimers. The overall fold is very similar to that of caspase-1 and caspase-3, but significant differences exist in the substrate-binding region. The structure answers questions about the enzyme-inhibitor complex that could not be explained from earlier caspase structures solved at lower resolution. The catalytic triad in caspase-8 comprises Cys360, His317 and the backbone carbonyl oxygen atom of Arg258, which points towards the Nepsilon atom of His317. The oxygen atom attached to the tetrahedral carbon in the thiohemiacetal group of the inhibitor is hydrogen bonded to Ndelta of His317, and is not in a region characteristic of a classical 'oxyanion hole'. The N-acetyl group of the inhibitor is in the trans configuration. The caspase-8-inhibitor structure provides the basis for understanding structure/function relationships in this important initiator of the proteolytic cascade that leads to programmed cell death.
- Structural, Analytical & Medicinal Chemistry Pharmacia & Upjohn Inc. 301 Henrietta Street, Kalamazoo, MI 49007, USA.
Organizational Affiliation: