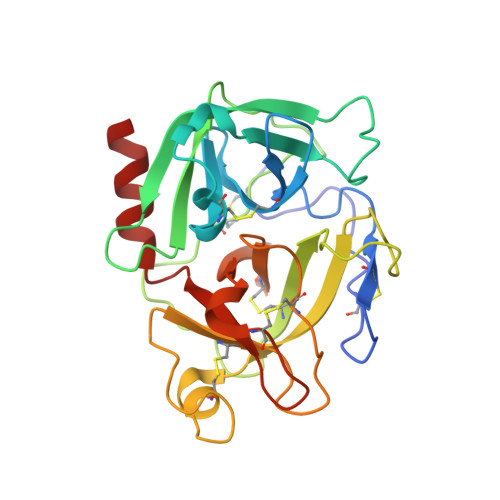
Crystal structure of the proenzyme domain of plasminogen.
Peisach, E., Wang, J., de los Santos, T., Reich, E., Ringe, D.(1999) Biochemistry 38: 11180-11188
- PubMed: 10460175
- DOI: https://doi.org/10.1021/bi991130r
- Primary Citation of Related Structures:
1QRZ - PubMed Abstract:
We have solved the X-ray crystal structure of the proenzyme form of the catalytic domain of plasminogen, with the nonessential mutations M585Q, V673M, and M788L, to 2.0 A resolution. The structure presents an inactive protease characterized by Asp740 (chymotrypsinogen 194) hydrogen bonded to His586 (chymotrypsinogen 40), preventing proper formation of the oxyanion hole and S1 specificity pocket. In addition, the catalytic triad residues are misplaced relative to the active conformation adopted by serine proteases in the chymotrypsin family. Finally, a unique form of zymogen inactivation is observed, characterized by a "foot-in-mouth" mechanism in which Trp761 (chymotrypsinogen 215) is folded into the S1 specificity pocket preventing substrate binding.
- Program in Biophysics and Structural Biology, Brandeis University, Waltham, Massachusetts 02454-9110, USA.
Organizational Affiliation: