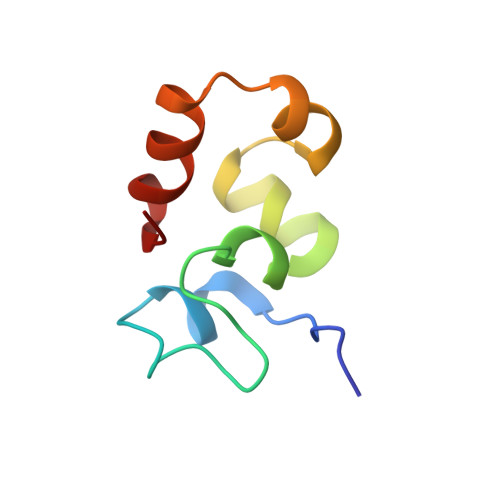
NMR structure of an F-actin-binding "headpiece" motif from villin.
Vardar, D., Buckley, D.A., Frank, B.S., McKnight, C.J.(1999) J Mol Biology 294: 1299-1310
- PubMed: 10600386
- DOI: https://doi.org/10.1006/jmbi.1999.3321
- Primary Citation of Related Structures:
1QQV - PubMed Abstract:
A growing family of F-actin-bundling proteins harbors a modular F-actin-binding headpiece domain at the C terminus. Headpiece provides one of the two F-actin-binding sites essential for filament bundling. Here, we report the first structure of a functional headpiece domain. The NMR structure of chicken villin headpiece (HP67) reveals two subdomains that share a tightly packed hydrophobic core. The N-terminal subdomain contains bends, turns, and a four-residue alpha-helix as well as a buried histidine residue that imparts a pH-dependent folding. The C-terminal subdomain is composed of three alpha-helices and its folding is pH-independent. Two residues previously implicated in F-actin-binding form a buried salt-bridge between the N and C-terminal subdomains. The rest of the identified actin-binding residues are solvent-exposed and map onto a unique F-actin-binding surface.
- Department of Biophysics, Boston University School of Medicine, 700 Albany Street, Boston, MA 02118, USA.
Organizational Affiliation: