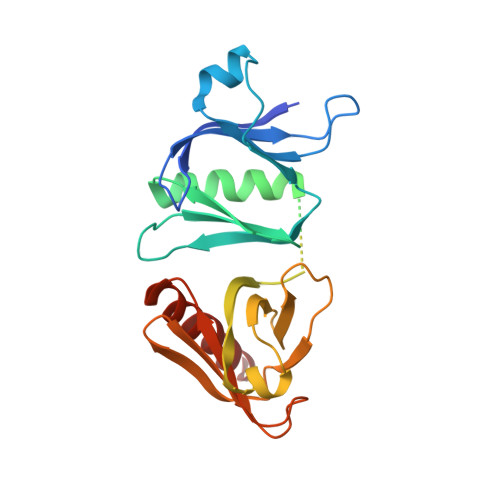
Crystal structure of the pleckstrin homology-phosphotyrosine binding (PH-PTB) targeting region of insulin receptor substrate 1.
Dhe-Paganon, S., Ottinger, E.A., Nolte, R.T., Eck, M.J., Shoelson, S.E.(1999) Proc Natl Acad Sci U S A 96: 8378-8383
- PubMed: 10411883
- DOI: https://doi.org/10.1073/pnas.96.15.8378
- Primary Citation of Related Structures:
1QQG - PubMed Abstract:
We have determined the crystal structure at 2.3-A resolution of an amino-terminal segment of human insulin receptor substrate 1 that encompasses its pleckstrin homology (PH) and phosphotyrosine binding (PTB) domains. Both domains adopt the canonical seven-stranded beta-sandwich PH domain fold. The domains are closely associated, with a 720-A(2) contact surface buried between them that appears to be stabilized by ionic, hydrophobic, and hydrogen bonding interactions. The nonconserved 46-residue linker between the domains is disordered. The PTB domain peptide binding site is fully exposed on the molecular surface, as is a large cationic patch at the base of the PH domain that is a likely binding site for the head groups of phosphatidylinositol phosphates. Binding assays confirm that phosphatidylinositol phosphates bind the PH domain, but not the PTB domain. Ligand binding to the PH domain does not alter PTB domain interactions, and vice versa. The structural and accompanying functional data illustrate how the two binding domains might act cooperatively to effectively increase local insulin receptor substrate 1 concentration at the membrane and transiently fix the receptor and substrate, to allow multiple phosphorylation reactions to occur during each union.
- Joslin Diabetes Center and the Department of Medicine, Harvard Medical School, Boston, MA 02215, USA.
Organizational Affiliation: