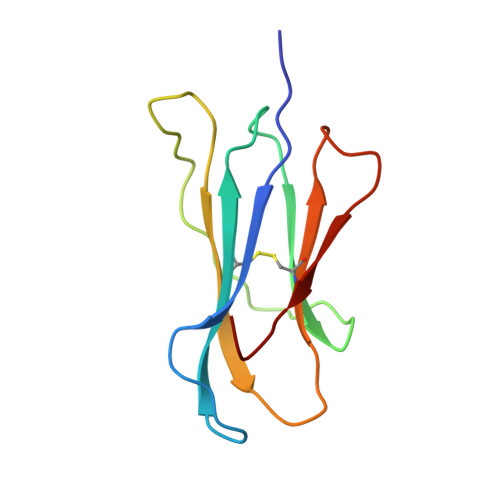
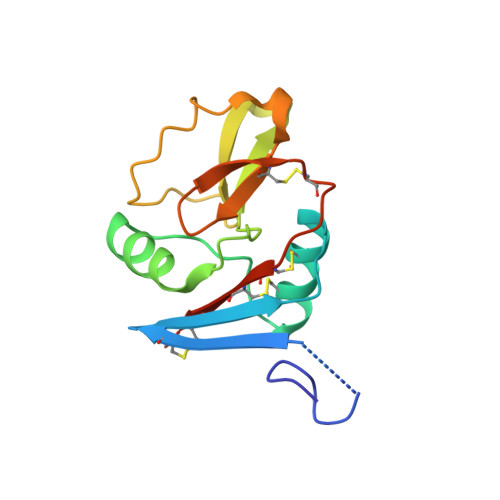
Crystal Structure of a Lectin-Like Natural Killer Cell Receptor Bound to its Mhc Class I Ligand
Tormo, J., Natarajan, K., Margulies, D.H., Mariuzza, R.A.(1999) Nature 402: 623
- PubMed: 10604468
- DOI: https://doi.org/10.1038/45170
- Primary Citation of Related Structures:
1QO3 - PubMed Abstract:
Natural killer (NK) cell function is regulated by NK receptors that interact with MHC class I (MHC-I) molecules on target cells. The murine NK receptor Ly49A inhibits NK cell activity by interacting with H-2D(d) through its C-type-lectin-like NK receptor domain. Here we report the crystal structure of the complex between the Ly49A NK receptor domain and unglycosylated H-2D(d). The Ly49A dimer interacts extensively with two H-2D(d) molecules at distinct sites. At one interface, a single Ly49A subunit contacts one side of the MHC-I peptide-binding platform, presenting an open cavity towards the conserved glycosylation site on the H-2D(d) alpha2 domain. At a second, larger interface, the Ly49A dimer binds in a region overlapping the CD8-binding site. The smaller interface probably represents the interaction between Ly49A on the NK cell and MHC-I on the target cell, whereas the larger one suggests an interaction between Ly49A and MHC-I on the NK cell itself. Both Ly49A binding sites on MHC-I are spatially distinct from that of the T-cell receptor.
- Center for Advanced Research in Biotechnology, University of Maryland Biotechnology Institute, Rockville 20850, USA.
Organizational Affiliation: