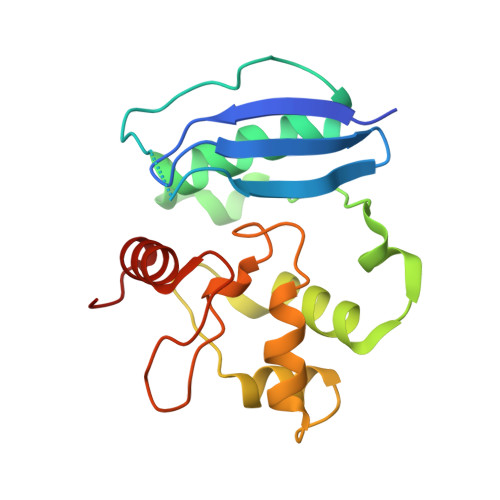
Crystal Structure of the Human O(6)-Alkylguanine-DNA Alkyltransferase.
Wibley, J.E.A., Pegg, A.E., Moody, P.C.E.(2000) Nucleic Acids Res 28: 393
- PubMed: 10606635
- DOI: https://doi.org/10.1093/nar/28.2.393
- Primary Citation of Related Structures:
1QNT - PubMed Abstract:
The mutagenic and carcinogenic effects of simple alkylating agents are mainly due to O(6)-alkylation of guanine in DNA. This lesion results in transition mutations. In both prokaryotic and eukaryotic cells, repair is effected by direct reversal of the damage by a suicide protein, O(6)-alkylguanine-DNA alkyltransferase. The alkyltransferase removes the alkyl group to one of its own cysteine residues. However, this mechanism for preserving genomic integrity limits the effectiveness of certain alkylating anticancer agents. A high level of the alkyltransferase in many tumour cells renders them resistant to such drugs. Here we report the X-ray structure of the human alkyltransferase solved using the technique of multiple wavelength anomalous dispersion. This structure explains the markedly different specificities towards various O(6)-alkyl lesions and inhibitors when compared with the Escherichia coli protein (for which the structure has already been determined). It is also used to interpret the behaviour of certain mutant alkyltransferases to enhance biochemical understanding of the protein. Further examination of the various models proposed for DNA binding is also permitted. This structure may be useful for the design and refinement of drugs as chemoenhancers of alkylating agent chemotherapy.
- Department of Biochemistry, University of Leicester, University Road, Leicester LE1 7RH, UK.
Organizational Affiliation: