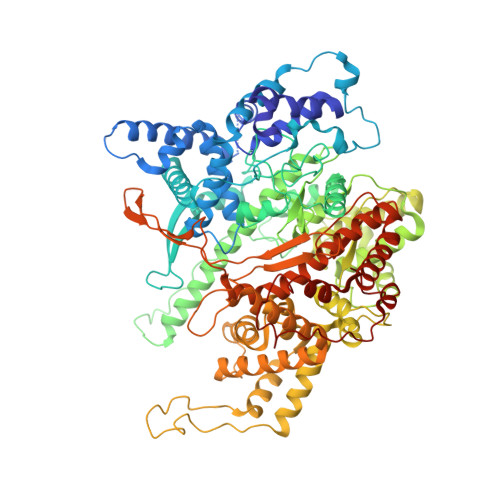
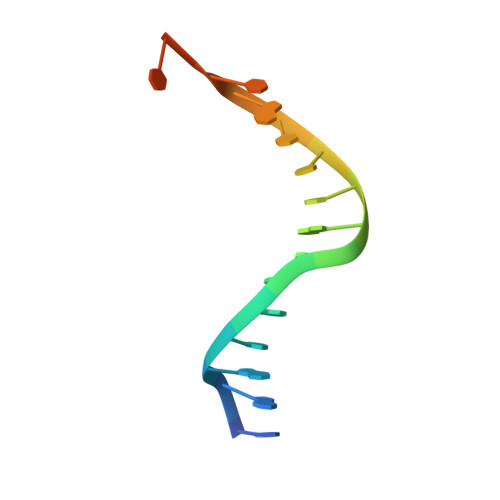
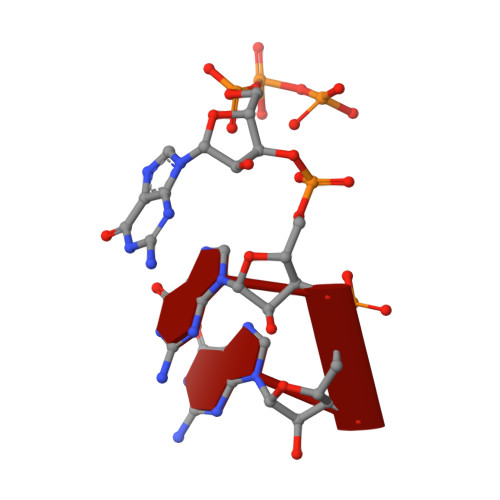
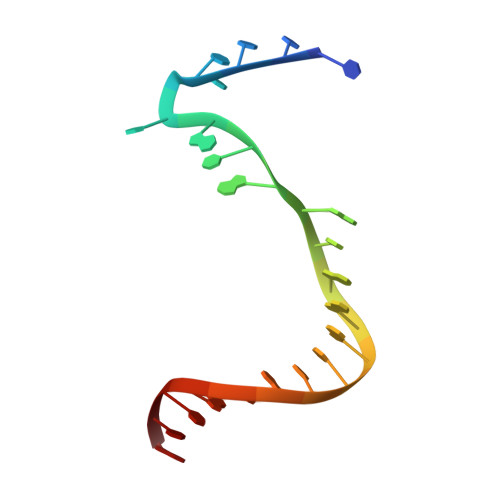
Structure of a Transcribing T7 RNA Polymerase Initiation Complex
Cheetham, G.M.T., Steitz, T.A.(1999) Science 286: 2305
- PubMed: 10600732
- DOI: https://doi.org/10.1126/science.286.5448.2305
- Primary Citation of Related Structures:
1QLN - PubMed Abstract:
The structure of a T7 RNA polymerase (T7 RNAP) initiation complex captured transcribing a trinucleotide of RNA from a 17-base pair promoter DNA containing a 5-nucleotide single-strand template extension was determined at a resolution of 2.4 angstroms. Binding of the upstream duplex portion of the promoter occurs in the same manner as that in the open promoter complex, but the single-stranded template is repositioned to place the +4 base at the catalytic active site. Thus, synthesis of RNA in the initiation phase leads to accumulation or "scrunching" of the template in the enclosed active site pocket of T7 RNAP. Only three base pairs of heteroduplex are formed before the RNA peels off the template.
- Department of Molecular Biophysics and Biochemistry, Yale University, Howard Hughes Medical Institute, New Haven, CT 06520-8114, USA.
Organizational Affiliation: