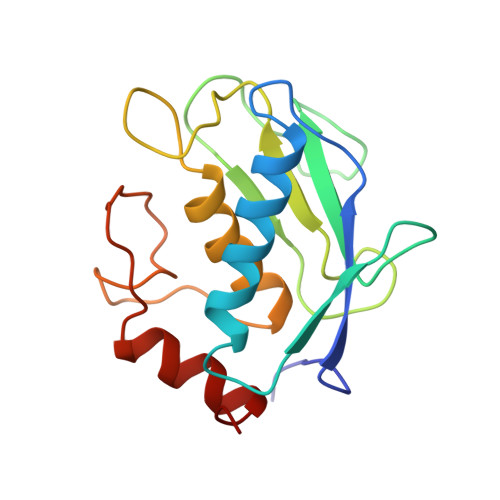
X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor
Dhanaraj, V., Williams, M.G., Ye, Q.-Z., Molina, F., Johnson, L.L., Ortwine, D.F., Pavlovsky, A., Rubin, J.R., Skeean, R.W., White, A.D., Humblet, C., Hupe, D.J., Blundell, T.L.(1999) Croat Chem Acta 72: 575-591