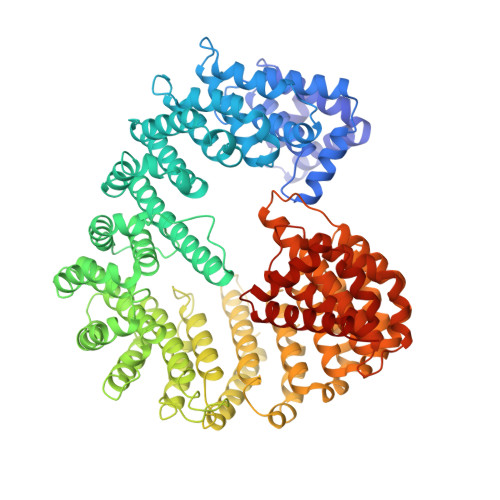
Structure of importin-beta bound to the IBB domain of importin-alpha.
Cingolani, G., Petosa, C., Weis, K., Muller, C.W.(1999) Nature 399: 221-229
- PubMed: 10353244
- DOI: https://doi.org/10.1038/20367
- Primary Citation of Related Structures:
1QGK, 1QGR - PubMed Abstract:
Cytosolic proteins bearing a classical nuclear localization signal enter the nucleus bound to a heterodimer of importin-alpha and importin-beta (also called karyopherin-alpha and -beta). The formation of this heterodimer involves the importin-beta-binding (IBB) domain of importin-alpha, a highly basic amino-terminal region of roughly 40 amino-acid residues. Here we report the crystal structure of human importin-beta bound to the IBB domain of importin-alpha, determined at 2.5 A and 2.3 A resolution in two crystal forms. Importin-beta consists of 19 tandemly repeated HEAT motifs and wraps intimately around the IBB domain. The association involves two separate regions of importin-beta, recognizing structurally distinct parts of the IBB domain: an amino-terminal extended moiety and a carboxy-terminal helix. The structure indicates that significant conformational changes occur when importin-beta binds or releases the IBB domain domain and suggests how dissociation of the importin-alpha/beta heterodimer may be achieved upon nuclear entry.
- European Molecular Biology Laboratory, Grenoble Outstation, France.
Organizational Affiliation: