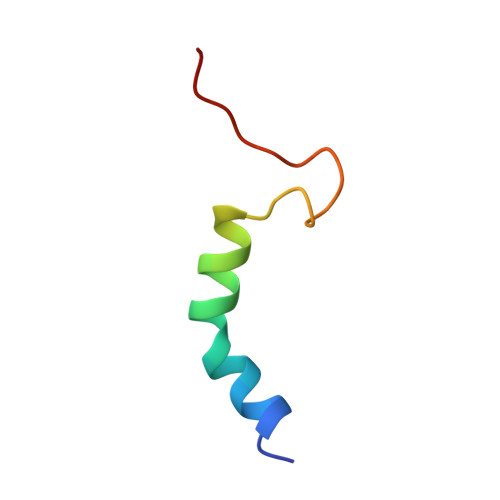
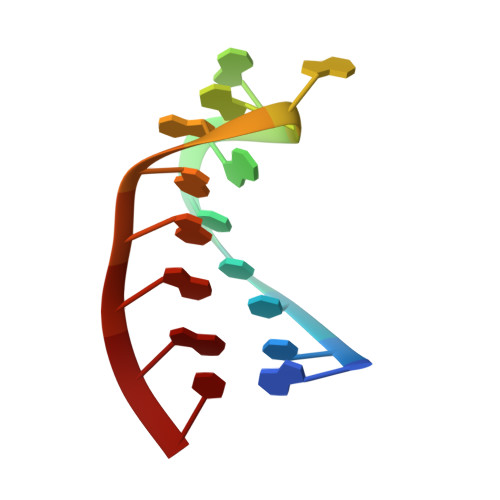
Antitermination in bacteriophage lambda. The structure of the N36 peptide-boxB RNA complex
Schaerpf, M., Sticht, H., Schweimer, K., Boehm, M., Hoffmann, S., Roesch, P.(2000) Eur J Biochem 267: 2397-2408
- PubMed: 10759866
- DOI: https://doi.org/10.1046/j.1432-1327.2000.01251.x
- Primary Citation of Related Structures:
1QFQ - PubMed Abstract:
The solution structure of a 15-mer nutRboxB RNA hairpin complexed with the 36-mer N-terminal peptide of the N protein (N36) from bacteriophage lambda was determined by 2D and 3D homonuclear and heteronuclear magnetic resonance spectroscopy. These 36 amino acids include the arginine-rich motif of the N protein involved in transcriptional antitermination of phage lambda. Upon complex formation with boxB RNA, the synthetic N36 peptide binds tightly to the major groove of the boxB hairpin through hydrophobic and electrostatic interactions forming a bent alpha helix. Four nucleotides of the GAAAA pentaloop of the boxB RNA adopt a GNRA-like tetraloop fold in the complex. The formation of a GAAA tetraloop involves a loop-closing sheared base pair (G6-A10), base stacking of three adenines (A7, A8, and A10), and extrusion of one nucleotide (A9) from the loop, as observed previously for the complex of N(1-22) peptide and the nutLboxB RNA [Legault, P., Li, J., Mogridge, J., Kay, L.E. & Greenblatt, J. (1998) Cell 93, 289-299]. Stacking of the bases is extended by the indole-ring of Trp18 which also forms hydrophobic contacts to the side-chains of Leu24, Leu25, and Val26. Based on the structure of the complex, three mutant peptides were synthesized and investigated by CD and NMR spectroscopy in order to determine the role of particular residues for complex formation. These studies revealed very distinct amino-acid requirements at positions 3, 4, and 8, while replacement of Trp18 with tyrosine did not result in any gross structural changes.
- Lehrstuhl für Biopolymere der Universität Bayreuth, Germany.
Organizational Affiliation: