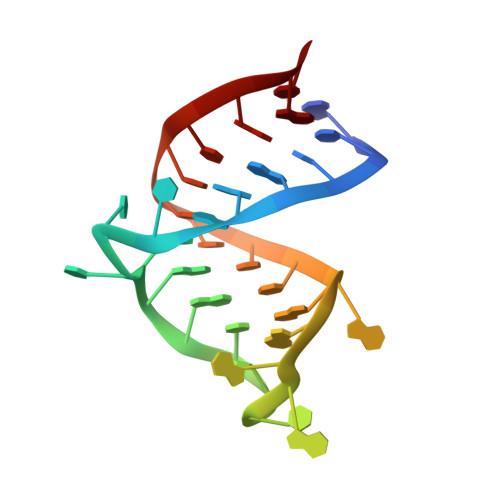
Structural rearrangements of HIV-1 Tat-responsive RNA upon binding of neomycin B.
Faber, C., Sticht, H., Schweimer, K., Rosch, P.(2000) J Biological Chem 275: 20660-20666
- PubMed: 10747964
- DOI: https://doi.org/10.1074/jbc.M000920200
- Primary Citation of Related Structures:
1QD3 - PubMed Abstract:
Binding of human immunodeficiency virus type 1 (HIV-1) transactivator (Tat) protein to Tat-responsive RNA (TAR) is essential for viral replication and is considered a promising starting point for the design of anti-HIV drugs. NMR spectroscopy indicated that the aminoglycosides neomycin B and ribostamycin bind to TAR and that neomycin is able to inhibit Tat binding to TAR. The solution structure of the neomycin-bound TAR has been determined by NMR spectroscopy. Chemical shift mapping and intermolecular nuclear Overhauser effects define the binding region of the aminoglycosides on TAR and give strong evidence for minor groove binding. Based on 15 nuclear Overhauser effect-derived intermolecular distance restraints, a model structure of the TAR-neomycin complex was calculated. Neomycin is bound in a binding pocket formed by the minor groove of the lower stem and the uridine-rich bulge of TAR, which adopts a conformation different from those known. The neamine core of the aminoglycoside (rings I and II) is covered with the bulge, explaining the inhibition of Tat by an allosteric mechanism. Neomycin reduces the volume of the major groove in which Tat is bound and thus impedes essential protein-RNA contacts.
- Lehrstuhl für Biopolymere, Universität Bayreuth, D-95447 Bayreuth, Germany.
Organizational Affiliation: