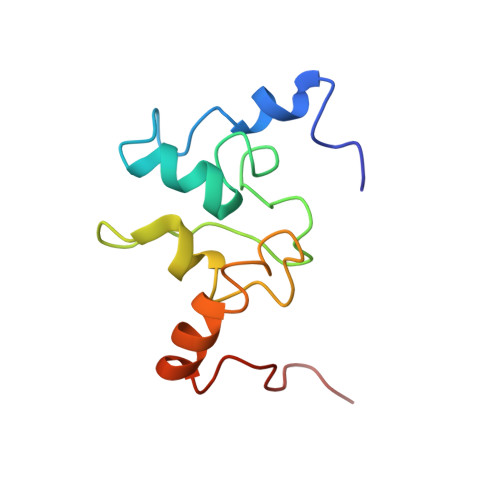
Solution structure of a baculoviral inhibitor of apoptosis (IAP) repeat.
Hinds, M.G., Norton, R.S., Vaux, D.L., Day, C.L.(1999) Nat Struct Biol 6: 648-651
- PubMed: 10404221
- DOI: https://doi.org/10.1038/10701
- Primary Citation of Related Structures:
1QBH - PubMed Abstract:
Members of the inhibitor of apoptosis (IAP) family of proteins are able to inhibit cell death following viral infection, during development or in cell lines in vitro. All IAP proteins bear one or more baculoviral IAP repeats (BIRs). Here we describe the solution structure of the third BIR domain from the mammalian IAP homolog B (MIHB/c-IAP-1). The BIR domain has a novel fold that is stabilized by zinc tetrahedrally coordinated by one histidine and three cysteine residues. The structure consists of a series of short alpha-helices and turns with the zinc packed in an unusually hydrophobic environment created by residues that are highly conserved among all BIRs.
- Biomolecular Research Institute, Parkville, Australia.
Organizational Affiliation: