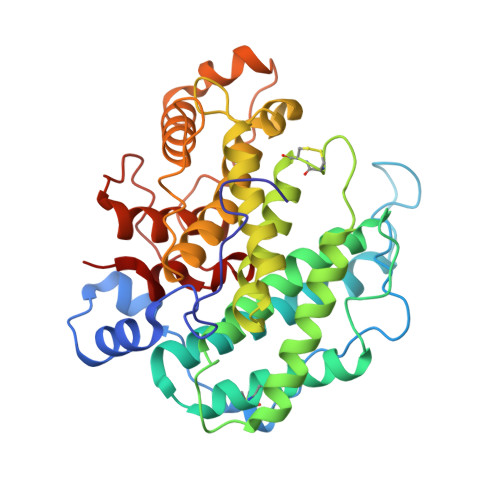
Crystal structure of alginate lyase A1-III from Sphingomonas species A1 at 1.78 A resolution.
Yoon, H.J., Mikami, B., Hashimoto, W., Murata, K.(1999) J Mol Biology 290: 505-514
- PubMed: 10390348
- DOI: https://doi.org/10.1006/jmbi.1999.2883
- Primary Citation of Related Structures:
1QAZ - PubMed Abstract:
The three-dimensional structure of alginate lyase A1-III (ALYIII) from a Sphingomonas species A1 was determined by X-ray crystallography. The enzyme was crystallized by the hanging-drop vapour-diffusion method in the presence of 49% ammonium sulfate at 20 degrees C. The crystals are monoclinic and belong to the space group C2 with unit cell dimensions of a=49.18 A, b=93.08 A, c=82.10 A and beta=104.12 degrees. There was one molecule of alginate lyase in the asymmetric unit of the crystal. The diffraction data up to 1. 71 A were collected with Rsymof 5.0%. The crystal structure of ALYIII was solved by the multiple isomorphous replacement method and refined at 1.78 A resolution using X-PLOR with a final R -factor of 18.0% for 10.0 to 1.78 A resolution data. The refined model of ALYIII contained 351 amino acid residues, 299 water molecules and two sulfate ions. The three-dimensional structure of ALYIII was abundant in helices and had a deep tunnel-like cleft in a novel (alpha6/alpha5)-barrel structure, which was similar to the (alpha6/alpha6)-barrel found in glucoamylase and cellulase. This structure presented the possibility that alginate molecules might penetrate into the cleft to interact with the catalytic site of ALYIII.
- Kyoto University, Uji Kyoto, 611-0011, Japan.
Organizational Affiliation: