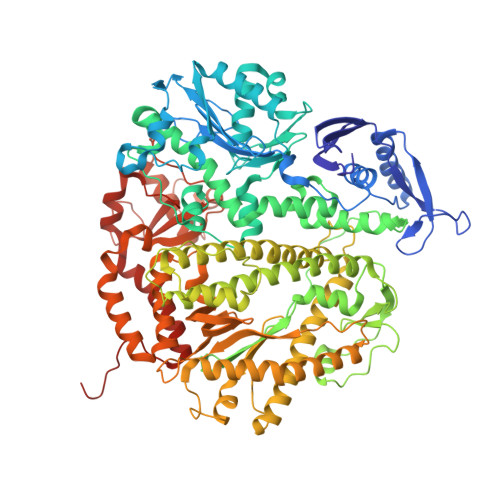
Lesion (in)tolerance reveals insights into DNA replication fidelity.
Freisinger, E., Grollman, A.P., Miller, H., Kisker, C.(2004) EMBO J 23: 1494-1505
- PubMed: 15057282
- DOI: https://doi.org/10.1038/sj.emboj.7600158
- Primary Citation of Related Structures:
1Q9X, 1Q9Y - PubMed Abstract:
The initial encounter of an unrepaired DNA lesion is likely to be with a replicative DNA polymerase, and the outcome of this event determines whether an error-prone or error-free damage avoidance pathway is taken. To understand the atomic details of this critical encounter, we have determined the crystal structures of the pol alpha family RB69 DNA polymerase with DNA containing the two most prevalent, spontaneously generated premutagenic lesions, an abasic site and 2'-deoxy-7,8-dihydro-8-oxoguanosine (8-oxodG). Identification of the interactions between these damaged nucleotides and the active site provides insight into the capacity of the polymerase to incorporate a base opposite the lesion. A novel open, catalytically inactive conformation of the DNA polymerase has been identified in the complex with a primed abasic site template. This structure provides the first molecular characterization of the DNA synthesis barrier caused by an abasic site and suggests a general mechanism for polymerase fidelity. In contrast, the structure of the ternary 8-oxodG:dCTP complex is almost identical to the replicating complex containing unmodified DNA, explaining the relative ease and fidelity by which this lesion is bypassed.
- Department of Pharmacological Sciences, Center for Structural Biology, SUNY at Stony Brook, Stony Brook, NY, USA.
Organizational Affiliation: