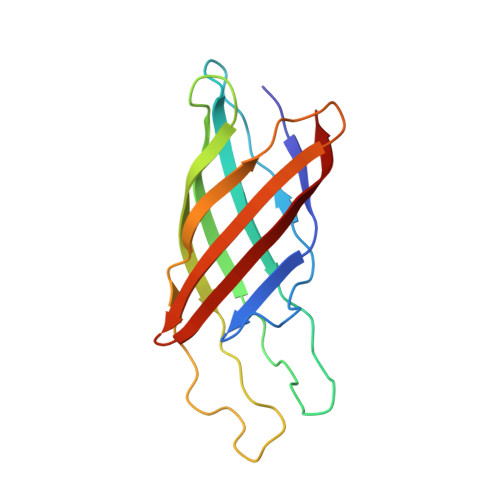
NMR structure of the integral membrane protein OmpX.
Fernandez, C., Hilty, C., Wider, G., Guntert, P., Wuthrich, K.(2004) J Mol Biology 336: 1211-1221
- PubMed: 15037080
- DOI: https://doi.org/10.1016/j.jmb.2003.09.014
- Primary Citation of Related Structures:
1Q9F, 1Q9G - PubMed Abstract:
The structure of the integral membrane protein OmpX from Escherichia coli reconstituted in 60 kDa DHPC micelles (OmpX/DHPC) was calculated from 526 NOE upper limit distance constraints. The structure determination was based on complete sequence-specific assignments for the amide protons and the Val, Leu, and Ile(delta1) methyl groups in OmpX, which were selectively protonated on a perdeuterated background. The solution structure of OmpX in the DHPC micelles consists of a well-defined, eight-stranded antiparallel beta-barrel, with successive pairs of beta-strands connected by mobile loops. Several long-range NOEs observed outside of the transmembrane barrel characterize an extension of a four-stranded beta-sheet beyond the height of the barrel. This protruding beta-sheet is believed to be involved in intermolecular interactions responsible for the biological functions of OmpX. The present approach for de novo structure determination should be quite widely applicable to membrane proteins reconstituted in mixed micelles with overall molecular masses up to about 100 kDa, and may also provide a platform for additional functional studies.
- Institut für Molekularbiologie und Biophysik, Eidgenössische Technische Hochschule Zürich CH-8093 Zürich, Switzerland.
Organizational Affiliation: