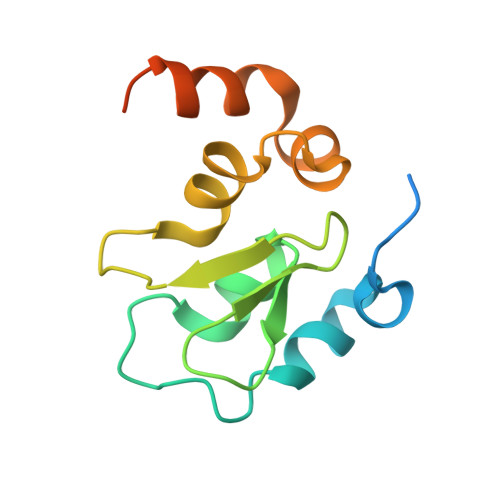
Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination
Chai, J., Yan, N., Huh, J.R., Wu, J.-W., Li, W., Hay, B.A., Shi, Y.(2003) Nat Struct Biol 10: 892-898
- PubMed: 14517550
- DOI: https://doi.org/10.1038/nsb989
- Primary Citation of Related Structures:
1Q4Q - PubMed Abstract:
The inhibitor of apoptosis protein DIAP1 inhibits Dronc-dependent cell death by ubiquitinating Dronc. The pro-death proteins Reaper, Hid and Grim (RHG) promote apoptosis by antagonizing DIAP1 function. Here we report the structural basis of Dronc recognition by DIAP1 as well as a novel mechanism by which the RHG proteins remove DIAP1-mediated downregulation of Dronc. Biochemical and structural analyses revealed that the second BIR (BIR2) domain of DIAP1 recognizes a 12-residue sequence in Dronc. This recognition is essential for DIAP1 binding to Dronc, and for targeting Dronc for ubiquitination. Notably, the Dronc-binding surface on BIR2 coincides with that required for binding to the N termini of the RHG proteins, which competitively eliminate DIAP1-mediated ubiquitination of Dronc. These observations reveal the molecular mechanisms of how DIAP1 recognizes Dronc, and more importantly, how the RHG proteins remove DIAP1-mediated ubiquitination of Dronc.
- Department of Molecular Biology, Princeton University, Lewis Thomas Laboratory, Washington Road, Princeton, New Jersey 08544, USA.
Organizational Affiliation: