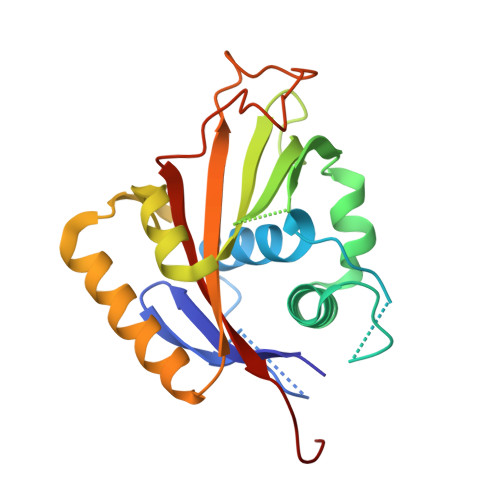
The Crystallographic Structure of Na,K-ATPase N-domain at 2.6 A Resolution
Hakansson, K.O.(2003) J Mol Biology 332: 1175-1182
- PubMed: 14499619
- DOI: https://doi.org/10.1016/j.jmb.2003.07.012
- Primary Citation of Related Structures:
1Q3I - PubMed Abstract:
The structure of the N-domain of porcine alpha(2) Na,K-ATPase was determined crystallographically to 3.2A resolution by isomorphous heavy-atom replacement using a single mercury derivative. The structure was finally refined against 2.6A resolution synchrotron data. The domain forms a seven-stranded antiparallel beta-sheet with two additional beta-strands forming a hairpin and five alpha-helices. Approximately 75% of the residues were superimposable with residues from the structure of Ca-ATPase N-domain, and a structure-based sequence alignment is presented. The positions of key residues are discussed in relation to the pattern of hydrophobicity, charge and sequence conservation of the molecular surface. The structure of a hexahistidine tag binding to nickel ions is presented.
- August Krogh Institute, Copenhagen University, Universitetsparken 13, DK-2100 OE, Copenhagen, Denmark. kohakansson@aki.ku.dk
Organizational Affiliation: