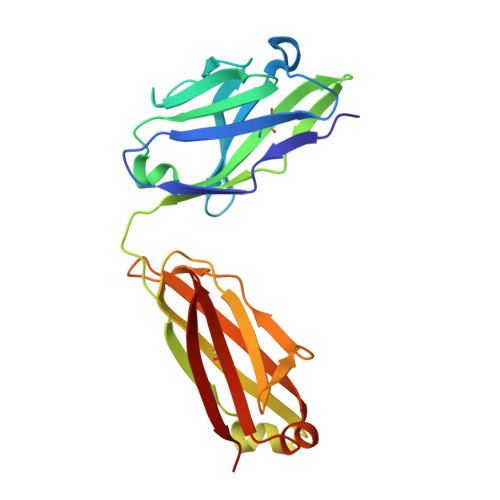
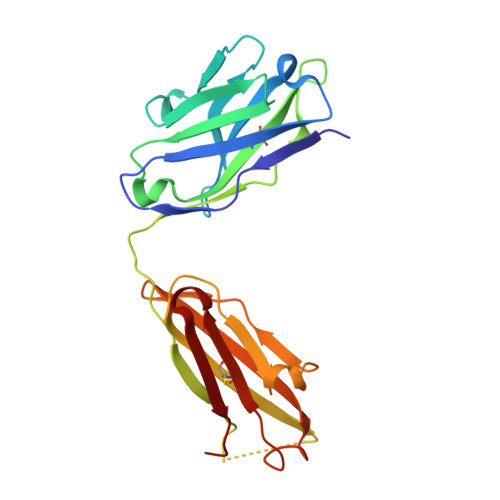
Anchoring a cationic ligand: the structure of the Fab fragment of the anti-morphine antibody 9B1 and its complex with morphine
Pozharski, E., Wilson, M.A., Hewagama, A., Shanafelt, A.B., Petsko, G., Ringe, D.(2004) J Mol Biology 337: 691-697
- PubMed: 15019787
- DOI: https://doi.org/10.1016/j.jmb.2003.12.084
- Primary Citation of Related Structures:
1Q0X, 1Q0Y - PubMed Abstract:
The crystal structures of an anti-morphine antibody 9B1 (to 1.6A resolution) and its complex with morphine (to 2.0 A resolution) are reported. The morphine-binding site is described as a shallow depression on the protein surface, an unusual topology for a high-affinity ( Ka approximately 10(9) M(-1)) antibody against a small antigen. The polar part of the ligand is exposed to solvent, and the cationic nitrogen atom of the morphine molecule is anchored at the bottom of the binding site by a salt-bridge to a glutamate side-chain. Additional affinity is provided by a double cation-pi interaction with two tryptophan residues. Comparison of the morphine complex with the structure of the free Fab shows that a domain closure occurs upon binding of the ligand.
- Rosenstiel Basic Medical Sciences Research Center, Brandeis University, 415 South Street, Waltham, MA 02454, USA.
Organizational Affiliation: