The Staphostatin-Staphopain Complex: A FORWARD BINDING INHIBITOR IN COMPLEX WITH ITS TARGET CYSTEINE PROTEASE.
Filipek, R., Rzychon, M., Oleksy, A., Gruca, M., Dubin, A., Potempa, J., Bochtler, M.(2003) J Biological Chem 278: 40959-40966
- PubMed: 12874290
- DOI: https://doi.org/10.1074/jbc.M302926200
- Primary Citation of Related Structures:
1PXV - PubMed Abstract:
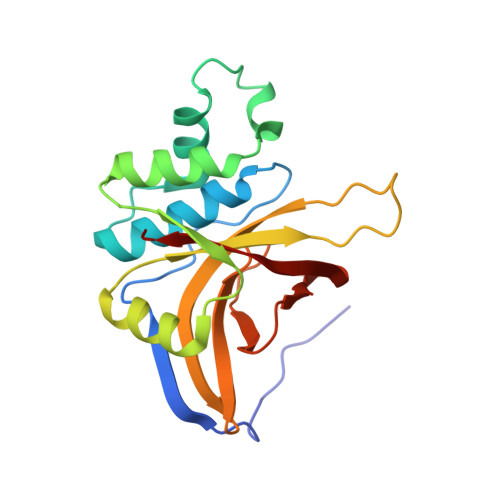
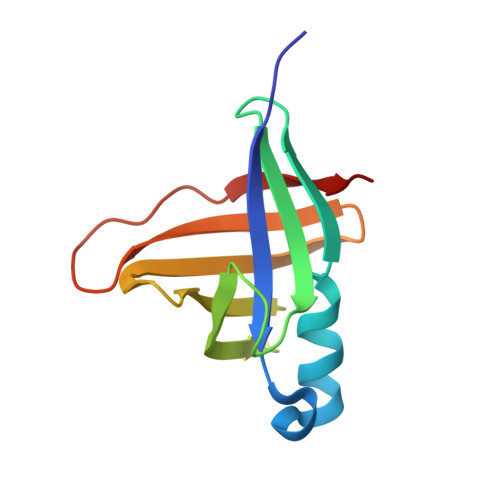
Staphostatins are the endogenous inhibitors of the major secreted cysteine proteases of Staphylococcus aureus, the staphopains. Our recent crystal structure of staphostatin B has shown that this inhibitor forms a mixed, eight-stranded beta-barrel with statistically significant similarity to lipocalins, but not to cystatins. We now present the 1.8-A crystal structure of staphostatin B in complex with an inactive mutant of its target protease. The complex is held together through extensive interactions and buries a total surface area of 2300 A2. Unexpectedly for a cysteine protease inhibitor, staphostatin B binds to staphopain B in an almost substrate-like manner. The inhibitor polypeptide chain runs through the protease active site cleft in the forward direction, with residues IG-TS in P2 to P2' positions. Both in the free and complexed forms, the P1 glycine residue of the inhibitor is in a main chain conformation only accessible to glycines. Mutations in this residue lead to a loss of affinity of the inhibitor for protease and convert the inhibitor into a substrate.
- International Institute of Molecular and Cell Biology, ul Trojdena 4, 02-109 Warsaw, Poland.
Organizational Affiliation: