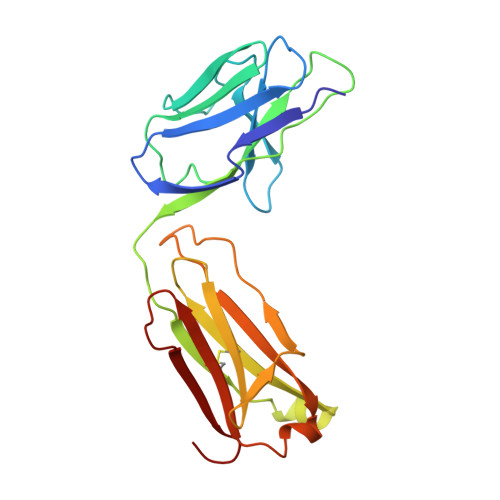
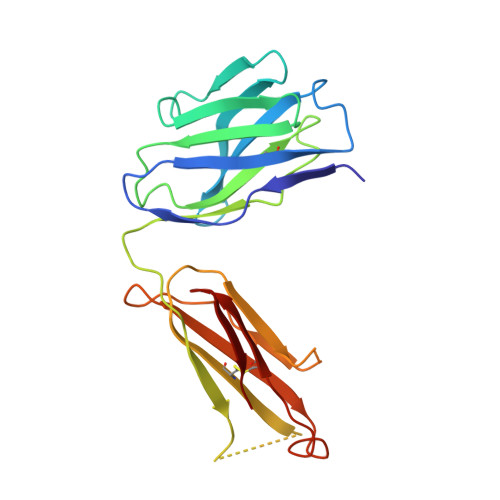
The crystal structure of a Fab fragment to the melanoma-associated GD2 ganglioside.
Pichla, S.L., Murali, R., Burnett, R.M.(1997) J Struct Biol 119: 6-16
- PubMed: 9216084
- DOI: https://doi.org/10.1006/jsbi.1997.3857
- Primary Citation of Related Structures:
1PSK - PubMed Abstract:
The GD2 ganglioside is a cell-surface component that appears on the surface of metastatic melanoma cells and is a marker for the progression of the disease. The ME36.1 monoclonal antibody binds to the GD2 ganglioside and has shown potential as a therapeutic antibody. ME36.1 is a possible alternative therapy to radiation, which is often ineffective in late-stage melanoma. The crystal structure of the Fab fragment of ME36.1 has been determined using molecular replacement and refined to an R factor of 20.4% at 2.8 A resolution. The model has good geometry with root-mean-square deviations of 0.008 A from ideal bond lengths and 1.7 degrees from ideal bond angles. The crystal structure of the ME36.1 Fab shows that its complementarity determining region forms a groove-shaped binding site rather than the pocket-type observed in other sugar binding Fabs. Molecular modeling has placed a four-residue sugar, representative of GD2, in the antigen binding site. The GD2 sugar moiety is stabilized by a network of hydrogen bonds that define the specificity of ME36.1 toward its antigen.
- Wistar Institute, Philadelphia, Pennsylvania 19104, USA.
Organizational Affiliation: