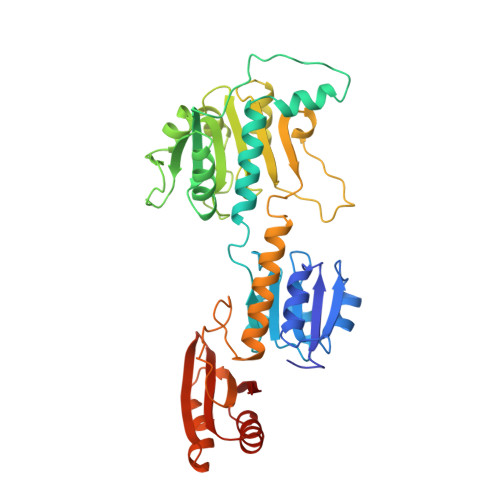
The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase.
Schuller, D.J., Grant, G.A., Banaszak, L.J.(1995) Nat Struct Biol 2: 69-76
- PubMed: 7719856
- DOI: https://doi.org/10.1038/nsb0195-69
- Primary Citation of Related Structures:
1PSD - PubMed Abstract:
The crystal structure of the phosphoglycerate dehydrogenase from Escherichia coli is unique among dehydrogenases. It consists of three clearly separate domains connected by flexible hinges. The tetramer has approximate 222 symmetry with the principal contacts between the subunits forming between either the nucleotide binding domains or the regulatory domains. Two slightly different subunit conformations are present which vary only in the orientations of the domains. There is a hinge-like arrangement near the active site cleft and the serine effector site is provided by the regulatory domain of each of two subunits. Interdomain flexibility may play a key role in both catalysis and allosteric inhibition.
- Department of Molecular Biology, University of California, Irvine 92717.
Organizational Affiliation: