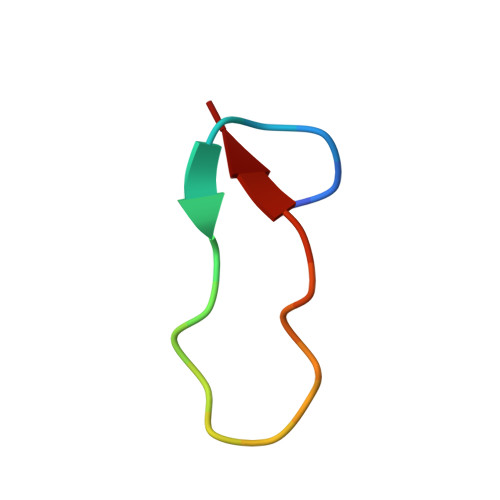
Structure of Antibacterial Peptide Microcin J25: A 21-Residue Lariat Protoknot.
Bayro, M.J., Mukhopadhyay, J., Swapna, G.V.T., Huang, J.Y., Ma, L.-C., Sineva, E., Dawson, P.E., Montelione, G.T., Ebright, R.H.(2003) J Am Chem Soc 125: 12382-12383
- PubMed: 14531661
- DOI: https://doi.org/10.1021/ja036677e
- Primary Citation of Related Structures:
1PP5 - PubMed Abstract:
The antibacterial peptide microcin J25 (MccJ25) inhibits bacterial transcription by binding within, and obstructing, the nucleotide-uptake channel of bacterial RNA polymerase. Published covalent and three-dimensional structures indicate that MccJ25 is a 21-residue cycle. Here, we show that the published covalent and three-dimensional structures are incorrect, and that MccJ25 in fact is a 21-residue "lariat protoknot", consisting of an 8-residue cyclic segment followed by a 13-residue linear segment that loops back and threads through the cyclic segment. MccJ25 is the first example of a lariat protoknot involving a backbone-side chain amide linkage.
- Department of Molecular Biology and Biochemistry, Waksman Institute, and Howard Hughes Medical Institute, Rutgers University, Piscataway, New Jersey 08854, USA.
Organizational Affiliation: