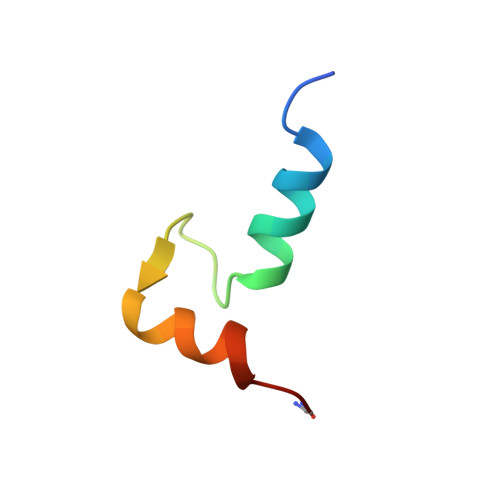
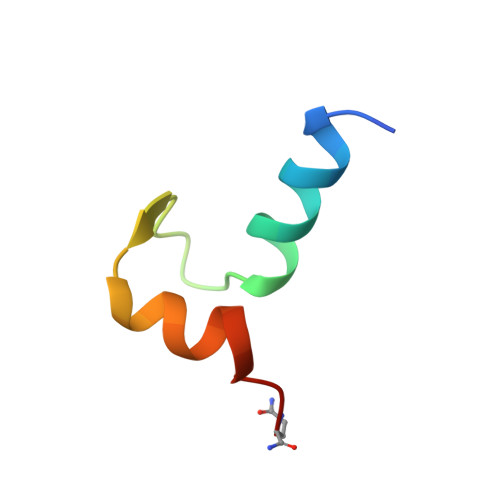
NMR solution structure of a synthetic troponin C heterodimeric domain.
Shaw, G.S., Sykes, B.D.(1996) Biochemistry 35: 7429-7438
- PubMed: 8652520
- DOI: https://doi.org/10.1021/bi9528006
- Primary Citation of Related Structures:
1PON - PubMed Abstract:
The C-terminal domain from the muscle protein troponin C (TnC) comprises two helix-loop-helix calcium-binding sites (residues 90-162). The assembly of these two sites is governed by calcium binding enabling a synthetic C-terminal domain to be preferentially and stoichiometrically assembled from two synthetic peptides (residues 93-126, SCIII, and 129-162, SCIV) in the presence of calcium only. It is therefore of great interest to know how closely the structure of this heterodimeric domain is to the intact protein domain. Analysis of such a structure has important implications in protein engineering and in understanding the stability of calcium-binding proteins in terms of biological function. The solution structure of this heterodimeric protein was determined by 1H NMR spectroscopy using 802 NOE derived distance restraints and 23 phi and 22 chi angle restraints. Distance geometry-simulated annealing calculations yielded a family of 42 converged structures (rmsd 0.86 +/- 0.17 A) showing an arrangement of four alpha-helices similar in fold to the C-terminal of troponin C. The dimer interface has several important interactions between helix pairs E/H and F/G responsible for the association of the two peptides. However, neither the peptide complex nor the solution NMR structure of TnC pack as tightly as that observed in the TnC X-ray structure. The interhelical distance between the F/G helix is about 1.4 A greater in solution than in the crystal. A comparison of the exposed surface area of the hydrophobic residues in the SCIII/SCIV heterodimer revealed that residues 1104, Y112, and 1121 are more exposed than in the previously determined solution structure of the SCIII homodimer. These residues are important for the interaction with the inhibitory region of TnI and provide evidence for their involvement in the regulation of muscle contraction.
- Department of Biochemistry & McLaughlin Macromolecular Structure Facility, University of Western Ontario, London, Canada.
Organizational Affiliation: