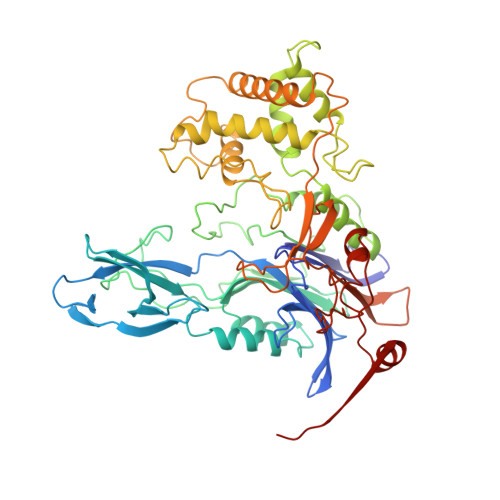
Penicillin acylase has a single-amino-acid catalytic centre.
Duggleby, H.J., Tolley, S.P., Hill, C.P., Dodson, E.J., Dodson, G., Moody, P.C.(1995) Nature 373: 264-268
- PubMed: 7816145
- DOI: https://doi.org/10.1038/373264a0
- Primary Citation of Related Structures:
1PNK, 1PNL, 1PNM - PubMed Abstract:
Penicillin acylase (penicillin amidohydrolase, EC 3.5.1.11) is widely distributed among microorganisms, including bacteria, yeast and filamentous fungi. It is used on an industrial scale for the production of 6-aminopenicillanic acid, the starting material for the synthesis of semi-synthetic penicillins. Its in vivo role remains unclear, however, and the observation that expression of the Escherichia coli enzyme in vivo is regulated by both temperature and phenylacetic acid has prompted speculation that the enzyme could be involved in the assimilation of aromatic compounds as carbon sources in the organism's free-living mode. The mature E. coli enzyme is a periplasmic 80K heterodimer of A and B chains (209 and 566 amino acids, respectively) synthesized as a single cytoplasmic precursor containing a 26-amino-acid signal sequence to direct export to the cytoplasm and a 54-amino-acid spacer between the A and B chains which may influence the final folding of the chains. The N-terminal serine of the B chain reacts with phenylmethylsulphonyl fluoride, which is consistent with a catalytic role for the serine hydroxyl group. Modifying this serine to a cysteine inactivates the enzyme, whereas threonine, arginine or glycine substitution prevents in vivo processing of the enzyme, indicating that this must be an important recognition site for cleavage. Here we report the crystal structure of penicillin acylase at 1.9 A resolution. Our analysis shows that the environment of the catalytically active N-terminal serine of the B chain contains no adjacent histidine equivalent to that found in the serine proteases. The nearest base to the hydroxyl of this serine is its own alpha-amino group, which may act by a new mechanism to endow the enzyme with its catalytic properties.
- Department of Chemistry, University of York, UK.
Organizational Affiliation: