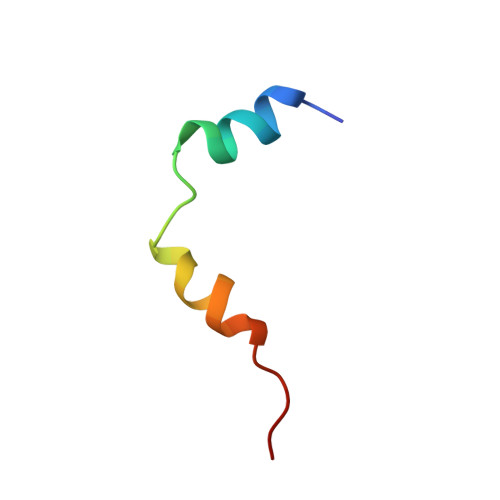
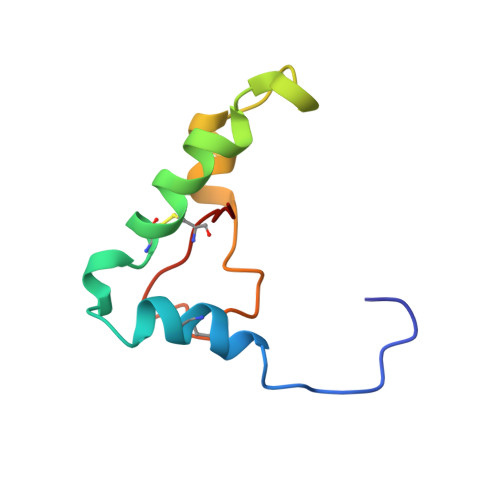
1H NMR assignment and global fold of napin BnIb, a representative 2S albumin seed protein.
Rico, M., Bruix, M., Gonzalez, C., Monsalve, R.I., Rodriguez, R.(1996) Biochemistry 35: 15672-15682
- PubMed: 8961930
- DOI: https://doi.org/10.1021/bi961748q
- Primary Citation of Related Structures:
1PNB - PubMed Abstract:
Napin BnIb is a representative member of the 2S albumin seed proteins, which consists of two polypeptide chains of 3.8 and 8.4 kDa linked by two disulfide bridges. In this work, a complete assignment of the 1H spectra of napin BnIb has been carried out by two-dimensional NMR sequence-specific methods and its secondary structure determined on the basis of spectral data. A calculation of the tertiary structure has been performed using approximately 500 distance constraints derived from unambiguously assigned NOE cross-correlations and distance geometry methods. The resulting global fold consists of five helices and a C-terminal loop arranged in a right-handed spiral. The folded protein is stabilized by two interchain disulfide bridges and two additional ones between cysteine residues in the large chain. The structure of napin BnIb represents a third example of a new and distinctive folding pattern first described for the hydrophobic protein from soybean and nonspecific lipid transfer proteins from wheat and maize. The presence of an internal cavity is not at all evident, which rules out in principle the napin BnIb as a carrier of lipids. The determined structure is compatible with activities attributed to these proteins such as phospholipid vesicle interaction, allergenicity, and calmodulin antagonism. Given the sequence homology of BnIb with other napins and napin-type 2S albumin seed proteins from different species, it is likely that all these proteins share a common architecture. The determined structure will be crucial to establish structure-function relationships and to explore the mechanisms of folding, processing, and deposition of these proteins. It will also provide a firm basis for a rational use of genetic engineering in order to develop improved transgenic plants.
- Instituto de Estructura de la Materia, CSIC, Madrid, Spain.
Organizational Affiliation: